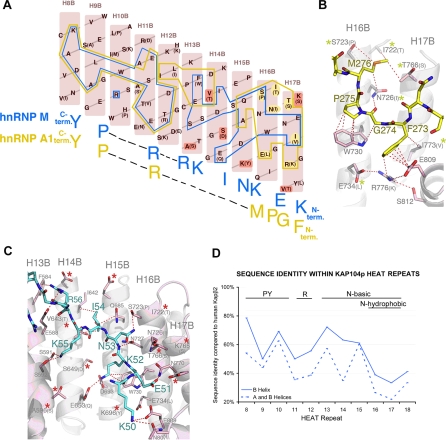
Figure 3. Kapβ2–NLS Structures and Kap104p Specificity.
(A) Schematic representation of the Kapβ2–NLS interface showing B helices of Kapβ2 H8–H17 (pink). Residues that are different in Kap104p are in parentheses. Kapβ2 residues that contact the hnRNP A1 NLS (hPY-NLS) and the hnRNP M NLS (bPY-NLS) are outlined in yellow and blue, respectively. Residues contacting the N-terminal FGPM hydrophobic motif in hnRNP A1, which are also different in Kap104p, are highlighted in yellow. Residues that increase electronegativity of the Kap104p surface are highlighted in red.
(B) Interactions between Kapβ2 (pink) and the N-terminal hydrophobic motif of hnRNP A1 (yellow) (PDB ID 2H4M) drawn with PYMOL [67]. Residues that are different in Kap104p are in parentheses (yellow asterisks label residues that may affect interactions with hPY-NLSs).
(C) Interactions between Kapβ2 (pink) and the N-terminal basic motif of hnRNP M (blue) (PDB ID 2OT8). Residues that are different in Kap104p are in parentheses. Red asterisks label Kap104p substitutions that increase electronegativity.
(D) Sequence identity within individual HEAT repeats of Kapβ2 and Kap104p. The motifs recognized by the B helix of each HEAT repeat are specified above the graph.