Fig. 2.
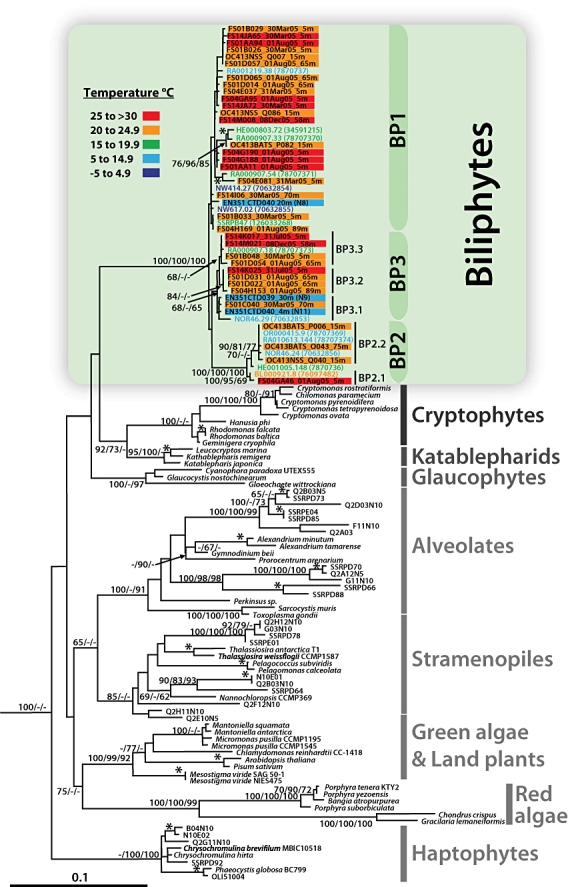
Phylogenetic analysis of 18S rRNA gene sequences from environmental clone libraries (all non-italic sequence names) and cultured representatives of eukaryotic first rank taxa. Sequences in coloured bars are reported for the first time in this study and are derived from 17 environmental clone libraries, three from the Sargasso Sea (prefixes: OC413BATS_P, Q; OC413NSS), 11 from three stations in the Florida Straits (prefixes: FS01, FS04, FS14) over multiple dates and depths (see Table 1), as well as three from continental shelf-edge and slope waters (prefixes: EN351). Biliphyte sequences from previous work are shown in coloured font. Bar and font colours correspond to five temperature ranges as indicated on the figure. Other Sargasso Sea and Pacific Ocean environmental sequences (non-italic, black font sequence names without coloured bars) are included to illustrate the distinct nature of biliphyte sequences relative to other uncultured eukaryotes retrieved from the same waters. The tree shown was inferred by maximum likelihood (ML) methods using the model TrN + I +Γ (I = 0.2332; Γ = 0.4748) with global rearrangements, randomized sequence input, 10 jumbles, Transition/Transversion = 1.9433 and six rate categories. Node values reflect bootstrap support in the order maximum likelihood/neighbour-joining distance/parsimony as percentages of 100/1000/100 replicates. Bootstrap support for terminal nodes (*) is indicated only if over 90% by all three methods. Two radiolarian sequences (126033309 and 126033226) served as an outgroup (not shown). The scale bar indicates the estimated number of nucleotide substitutions per site.
