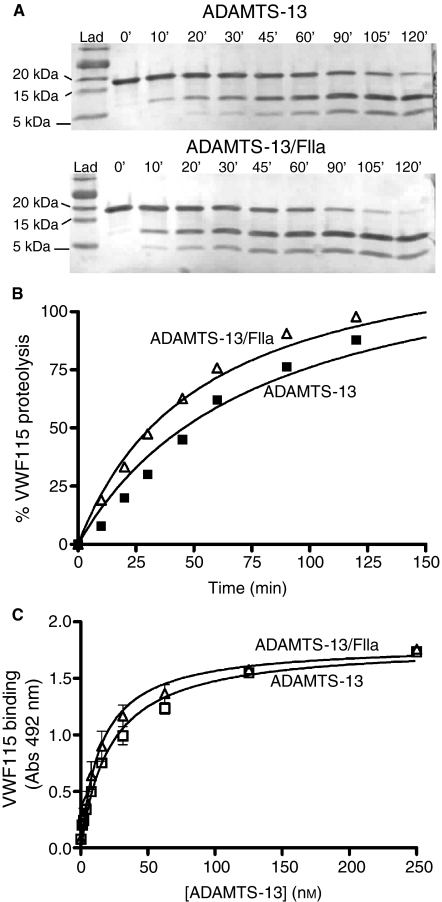
Fig. 4.
Proteolysis of ADAMTS-13 by thrombin does not influence the cleavage of, or binding to, VWF115. (A) 5 nm ADAMTS-13 or thrombin-cleaved ADAMTS-13 (ADAMTS-13/FIIa) was incubated at 37 °C with 5 μm VWF115, 150 mm NaCl, 5 mm CaCl2, 20 mm Tris (pH7.8). Subsamples were stopped with EDTA (0–2 h) and analyzed by sodium dodecylsulfate polyacrylamide gel electrophoresis and Coomassie staining. Proteolysis of VWF115 was visualized by the disappearance of a 16.9 kDa band and the appearance of 10 kDa and 6.9 kDa cleavage products. (B) Kinetic analysis of VWF115 proteolysis, as in (A) except using 700 nm VWF115. Proteolysis of VWF115 was quantified by high-performance liquid chromatography, and represented graphically. The catalytic efficiency for VWF115 proteolysis was derived from fitted data for both ADAMTS-13 and ADAMTS-13/FIIa (8 × 104 m−1 s−1 and 11 × 104 m−1 s−1, respectively). (C) The affinity of ADAMTS-13 and thrombin-cleaved ADAMTS-13 for VWF115 was measured by plate assay. 140 nm VWF115 was immobilized to microtiter wells, and incubated with varying concentrations of ADAMTS-13 or thrombin-cleaved ADAMTS-13 (ADAMTS-13/FIIa), as in Materials and methods. A similar high-affinity interaction was determined (KD∼20 nm) for both ADAMTS-13 and ADAMTS-13/FIIa with VWF115.