Abstract
Hepatitis B virus (HBV)-induced liver failure is an emergent liver disease leading to high mortality. The severity of liver failure may be reflected by the profile of some metabolites. This study assessed the potential of using metabolites as biomarkers for liver failure by identifying metabolites with good discriminative performance for its phenotype. The serum samples from 24 HBV-induced liver failure patients and 23 healthy volunteers were collected and analyzed by gas chromatography-mass spectrometry (GC-MS) to generate metabolite profiles. The 24 patients were further grouped into two classes according to the severity of liver failure. Twenty-five commensal peaks in all metabolite profiles were extracted, and the relative area values of these peaks were used as features for each sample. Three algorithms, F-test, k-nearest neighbor (KNN) and fuzzy support vector machine (FSVM) combined with exhaustive search (ES), were employed to identify a subset of metabolites (biomarkers) that best predict liver failure. Based on the achieved experimental dataset, 93.62% predictive accuracy by 6 features was selected with FSVM-ES and three key metabolites, glyceric acid, cis-aconitic acid and citric acid, are identified as potential diagnostic biomarkers.
Keywords: Metabolite profile analysis, Potential diagnostic biomarker identification, k-nearest neighbor (KNN), Fuzzy support vector machine (FSVM), Exhaustive search (ES), Gas chromatography-mass spectrometry (GC-MS), Hepatitis B virus (HBV)-induced liver failure
INTRODUCTION
It is estimated that more than 350 million people are infected with hepatitis B virus (HBV) globally, of which more than 120 million people are in China (Li et al., 2005). HBV infection is a major cause of many liver diseases, such as chronic/severe hepatitis, cirrhosis, hepatocellular carcinoma and liver failure, in which liver failure is the most emergent one leading to high mortality (Lee, 1997).
When one suffers from liver diseases, the content of metabolites, especially endogenous compounds with low and middle molecular weights in serum, may be different from their normal levels (McConnell et al., 1995), since the liver is the main organ for metabolism. Analysis of these metabolites is a promising way for understanding and elucidating the etiology and mechanism of liver diseases (Alaoui-Jamali and Xu, 2006; Nicholson et al., 1999; Yang et al., 2004).
Liver failure induces severe metabolic disturbance (Nicholson and Wilson, 2003), so comprehensive metabolite profiling may be helpful to recognize phenotypes of the disease and the profile analysis may be useful to identify the potential biochemical markers for prediction or prognosis of the disease progression. In this study, metabolite profiling by gas chromatography-mass spectrometry (GC-MS) was employed to investigate the metabolites in serum samples from 24 HBV-induced liver failure patients and 23 healthy volunteers. All samples are grouped into 3 groups, and 25 commensal features are extracted to characterize them. The potential of corresponding metabolites was evaluated by feature selection algorithms for multiclass dataset.
Feature selection methods can be categorized into two kinds: filter and wrapper methods (Inza et al., 2004). Filter method is independent of the classification algorithm. It ranks or scores each feature based on measures such as the information gain criterion, mutual information, relief, etc., and then finds out optimal feature subset according to heuristic rules made on these ranks or scores. Therefore, in many circumstances, the construction of filter method used in binary decision is similar to that used in multiclass decision. The main advantage of the filter method is its low computational cost due to never engaging with learning algorithm, and its weak point lies on that it is difficult to note the complicated relations among features and between features and classification labels (Luts et al., 2007). The optimal feature subset selected by filter method always has bigger sizes and weaker performances than that selected by the wrapper method that takes the learning algorithm into account. By wrapper method, different feature subsets are tried to combine with the classification algorithm to estimate their performance, and the best set is kept as the result. Many search strategies, such as exhaustive search, best first search, stochastic search, and heuristic search like backward, forward or stepwise, are often used to explore the feature subsets in feature combinational space (Inza et al., 2004). To evaluate each feature subset, n-fold cross-validation (CV) and leave-one-out (LOO) are usually employed. The main disadvantage of wrapper method is the high computational cost of searching for feature subset. Heuristic search is the fastest strategy used in wrapper method, but the limitation in its design lies on heuristic/indirect feature evaluation rule which is usually based on construction of specific decision algorithm. In multiclass decision algorithm, heuristic/indirect feature evaluation rule is difficult to be introduced due to complexity of multiclass decision function. Direct feature evaluation by CV or LOO is an effective method in feature selection for multiclass decision, and when combined with exhaustive search (ES), it will give out the best results than any other search strategy, because ES traverses the whole feature combination space (Theodoridis and Koutroumbas, 2003). CV/LOO combined with exhaustive feature subset search is a feasible means on dataset with small feature number, but the main problem lies on whether the choice of multiclass decision machine used in CV/LOO will influence the final feature selection results.
In this work, a filter method (F-test) and two new wrapper methods (k-nearest neighbor (KNN) combined with ES (KNN-ES) based on CV and fuzzy support vector machine (FSVM) (Mao et al., 2005) combined with ES (FSVM-ES) based on CV) were compared for potential diagnostic metabolite selection application. To accelerate these algorithms, feature pre-selection was done by t-test according to features’ P value between every two classes. Features with no significant difference between any two classes were excluded. KNN combined with CV and FSVM combined with CV were chosen as feature subset evaluation tools to describe the feature selection performance in the three methods.
MATERIALS AND METHODS
Collection of serum samples
Spontaneous blood samples were collected from 24 HBV-induced liver failure patients (21 males and 3 females) and 23 healthy volunteers (15 males, 8 females). The patients aged 33~60 years, and the healthy volunteers aged 18~36 years. The mean age of patients and healthy volunteers were 46 and 27 years, respectively. The MELD scores for patients were 16.3~33.1, and the medium value was 21.8 and the mean value was 24.7. The blood samples were centrifuged at 3000 r/min for 10 min at 4 °C, and the sera were stored at −80 °C until use.
Determination of different metabolites
All serum samples are treated by GC-MS experiments to achieve metabolite profiles. Each sample was represented with a GC-MS total ion current (TIC) chromatogram. After peak deconvolution, identification and matching, 25 commensal peaks in all metabolite profiles were extracted and the relative area values of these peaks were used as features to characterize each serum sample. The metabolites corresponding to these peaks were identified according to mass spectra library (version 1.0.0.12) of National Institute of Standards and Technology (NIST).
Data preprocessing
The 24 HBV-induced liver failure patients were grouped into two classes by MELD threshold value 22.0 according to their different severity degrees, which are between the mean value and the medium value of all patients’ MELD scores (Kamath et al., 2001). The group with smaller MELD scores consisted of 13 patients with mild liver failure, and the other group had 11 patients with severer liver failure. The 23 healthy volunteers were included into control group. To depress the influence of a quantitative level of every feature on the processing algorithm of following data, Z-score normalization across all samples was employed on all 25 features in the entire dataset. To accelerate feature selection procedure, features without discriminant ability were excluded from feature pre-selection procedures, in which a two-tailed t-test was used to calculate features’ P value between every two classes; features without significant difference between any two classes were excluded. A threshold value for P value is set, and if a feature’s all P values are all above the threshold, the corresponding feature will be eliminated.
Feature selection algorithms
1. F-test-based feature selection
F-test is a filter method to evaluate features’ importance by calculating a ranking score as:
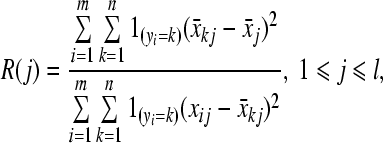 |
where j corresponds to a specific feature; i points to a specific sample; k indicates a specific class; m is total number of samples; n is total number of classes; l is total number of features; xij denotes the value of feature j in sample i; yi denotes the class label of sample i; x̄j denotes the mean value of feature j across all samples; x̄kj denotes the mean value of feature j across all samples belonging to class k; the indicator function 1Ω is equal to 1 if the event Ω is true and 0 otherwise. All features were ranked in a list according to their scores, and the combination of several top features with best discriminant performance and relatively small size was considered as target feature subset, and these top features are considered as potential diagnostic biomarkers.
2. KNN-ES algorithm
KNN-ES algorithm is a wrapper method, in which ES is employed to traverse all feature combinations to ensure discovery of globally optimal feature subset. KNN-based n-fold CV accuracy is the main evaluation index to estimate discriminative performance of all feature subsets. The confusion matrix is used as a second index to evaluate the accuracy and balance of classification error rates between different classes globally (Parker, 2001). KNN is a feasible nonparametric multi-classification method in supervised learning strategies. In KNN, the Euclidean distance between a specific sample and each of the other samples in training set is calculated, and the specific sample is classified to the class consisting of the most samples in its KNN. The only parameter in KNN is k. If a bigger k is used, the algorithm performs more robustly, but k should be smaller than the number of samples in the class consisting of the smallest samples in training set (Dasarathy, 1991). In Enas and Choi (1986)’s work, k=m
2/8 or k=m
3/8 is concluded as another rule to select k, where m is total number of samples. In our work, k is set according to these rules. By ES strategy, 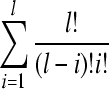 is explored, where l is total number of features. In so many feature subsets, the optimal feature subset is usually not exclusive. Many feature subsets will show similar top performance. If only features in optimal feature subset are thought as biomarkers, the results will be unstable. So feature frequency statistical histogram in a population of best feature subsets is introduced to evaluate the importance of features. The most popular features are thought as potential biomarkers. Using KNN-based n-fold CV accuracy as the main discriminative performance index, best feature subsets mean their predictive accuracy in excesses of a given threshold value.
is explored, where l is total number of features. In so many feature subsets, the optimal feature subset is usually not exclusive. Many feature subsets will show similar top performance. If only features in optimal feature subset are thought as biomarkers, the results will be unstable. So feature frequency statistical histogram in a population of best feature subsets is introduced to evaluate the importance of features. The most popular features are thought as potential biomarkers. Using KNN-based n-fold CV accuracy as the main discriminative performance index, best feature subsets mean their predictive accuracy in excesses of a given threshold value.
3. FSVM-ES algorithm
FSVM-ES algorithm is another wrapper method. Difference between FSVM-ES and KNN-ES is FSVM instead of KNN to be used to evaluate discriminative performance of feature subsets. FSVM is a relatively new multi-classification method (Mao et al., 2005; 2007) with remarkable robust performance on sparse and noisy data. In the process of training, if there are n classes in a training dataset, there are n(n−1)/2 support vector machines (SVMs) trained in FSVM. The decision function of FSVM is constructed based on these n(n−1)/2 SVM classification functions described as:
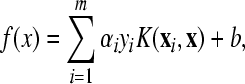 |
where the coefficients α=(αi) and b are obtained by training over a set of samples 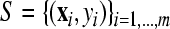 ;
;
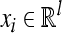 is sample i vector, and l is total number of features; yi is class label of sample i; K(·,·) is the kernel function; m is total number of samples (Vapnik, 1999). Define fst(x)=−fts(x) (s≠t; s, t=1, …, n). The fuzzy membership function gst(x) is introduced as:
is sample i vector, and l is total number of features; yi is class label of sample i; K(·,·) is the kernel function; m is total number of samples (Vapnik, 1999). Define fst(x)=−fts(x) (s≠t; s, t=1, …, n). The fuzzy membership function gst(x) is introduced as:
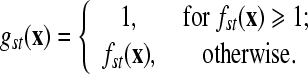 |
Using gst(x) (s≠t; s,t=1, …, n) and the class s (s∈{1, …, n}), the membership of x is defined as 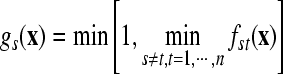 . An unknown sample x is classified by
. An unknown sample x is classified by 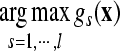 . In many circumstances, almost all samples in a dataset can be separated by linear decision function, so linear kernel FSVM was chosen as decision machine in our work, in which K(·,·) is defined as Euclidean inner product function. Another advantage of using linear kernel SVM is that only one parameter needs to be adjusted in the algorithm: the penalty parameter C, which is usually set as a constant in applications, e.g., 100 (Mao et al., 2005). Statistical histogram of selection frequency of each feature in a population of best feature subsets is also introduced to identify potential biomarkers. Here, the best feature subsets are determined by a given threshold on FSVM-based n-fold CV accuracy.
. In many circumstances, almost all samples in a dataset can be separated by linear decision function, so linear kernel FSVM was chosen as decision machine in our work, in which K(·,·) is defined as Euclidean inner product function. Another advantage of using linear kernel SVM is that only one parameter needs to be adjusted in the algorithm: the penalty parameter C, which is usually set as a constant in applications, e.g., 100 (Mao et al., 2005). Statistical histogram of selection frequency of each feature in a population of best feature subsets is also introduced to identify potential biomarkers. Here, the best feature subsets are determined by a given threshold on FSVM-based n-fold CV accuracy.
RESULTS AND DISCUSSION
Nineteen features were pre-selected by t-test with threshold value 0.05 for P value (Table 1). Based on the 3-class dataset with the 19 features, 5-fold CV accuracy was chosen as the main discriminative performance evaluation index for feature subsets. Confusion matrix was used as assisted index to evaluate the accuracy and balance of classification error rates among different classes globally, and contained discriminant state statistics between any two classes. In KNN-ES, k used in KNN may be set to 3 or 4 according to the rules mentioned in subsection “KNN-ES algorithm”, but no difference was found between the two corresponding results. We chose k=4 to elaborate the performance of KNN-ES. In FSVM-ES, penalty parameters were set to 100. All algorithms were implemented in Matlab codes.
Table 1.
List of endogenous serum metabolites measured by GC-MS
| Peak No. | Metabolites identified |
P value |
||
| Classes 1 vs 2 | Classes 1 vs 3 | Classes 2 vs 3 | ||
| 1 | Glycine | 0.0268 | 0.1643 | 0.0824 |
| 2 | L-threonine | 0.0038 | 0.0202 | 0.2882 |
| 3 | Glycine | 0.0385 | 0.0227 | 0.1606 |
| 4 | Glyceric acid | 0.6478 | 0.0418 | 0.0238 |
| 5 | L-serine | 0.0002 | 0.0000 | 0.0177 |
| 6 | L-threonine | 0.0015 | 0.0000 | 0.0100 |
| 7 | Proline | 0.9682 | 0.0032 | 0.0253 |
| 8 | L-proline | 0.0217 | 0.0000 | 0.0299 |
| 9 | L-serine | 0.2356 | 0.0140 | 0.0788 |
| 10 | L-proline | 0.0699 | 0.0015 | 0.0717 |
| 11 | Glutamine | 0.0798 | 0.0002 | 0.0077 |
| 12 | L-phenylalanine | 0.0033 | 0.0000 | 0.0145 |
| 13 | Ribitol | 0.8654 | 0.0434 | 0.1300 |
| 14 | cis-aconitic acid | 0.1927 | 0.0000 | 0.0016 |
| 15 | Citric acid | 0.0001 | 0.0003 | 0.0077 |
| 16 | 2-deoxy-galactopyranose | 0.0166 | 0.0232 | 0.8504 |
| 17 | D-fructose | 0.0002 | 0.3809 | 0.1869 |
| 18 | Hexadecanoic acid | 0.9249 | 0.0326 | 0.1109 |
| 19 | myo-inositol | 0.2567 | 0.0019 | 0.0418 |
| 20 | Ethanedioic acid | 0.2555 | 0.2024 | 0.0572 |
| 21 | L-valine | 0.2822 | 0.1352 | 0.1118 |
| 22 | D-ribose | 0.7668 | 0.0636 | 0.1485 |
| 23 | D-turanose | 0.2281 | 0.9954 | 0.6709 |
| 24 | D-glucose | 0.2084 | 0.4619 | 0.3811 |
| 25 | D-galactose | 0.2921 | 0.4521 | 0.3625 |
Note: Class 1 is normal group, Class 2 is mild liver failure group, and Class 3 is severe liver failure group. The peak numbers from 20 to 25 correspond to the metabolites excluded by t-test
Data analysis for single metabolite feature
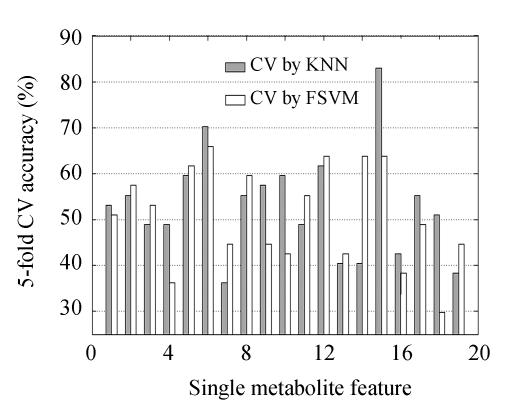
Using FSVM and KNN as classifiers, the 5-fold CV accuracy for each metabolite feature is described in Fig.1. Features 5, 6, 12 and 15 were identified by these two evaluation methods with discriminant accuracy ≥60%, and the t-test P scores of these four features between any two classes in Table 1 were all below 0.02. Using 5-fold CV by KNN, the discriminant accuracy for Feature 15 was 82.98%; and by FSVM-based 5-fold CV evaluation, its discriminant accuracy was 63.83%. In Fig.1, no feature shows remarkable linear discriminant power, and Feature 15 seems to be more discriminative than other features.
Fig. 1.
Discriminative performance evaluation of single metabolite feature by KNN/FSVM-based 5-fold CV accuracy
Biomarker selection by F-test statistics
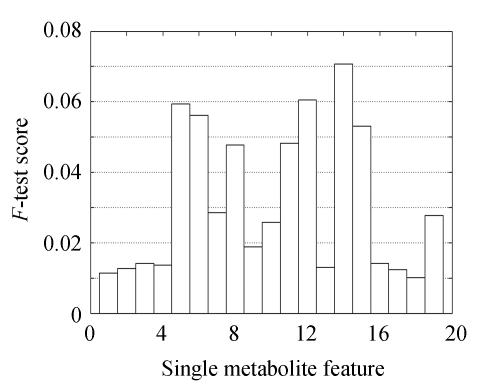
All features were ranked in a list according to their F-test scores, and features with maximal scores were ranked in top positions. These scores are described in Fig.2, in which Features 5, 6, 12, 14 and 15 were indicated as remarkable ones with scores greater than 0.05.
Fig. 2.
F-test score for each single metabolite feature
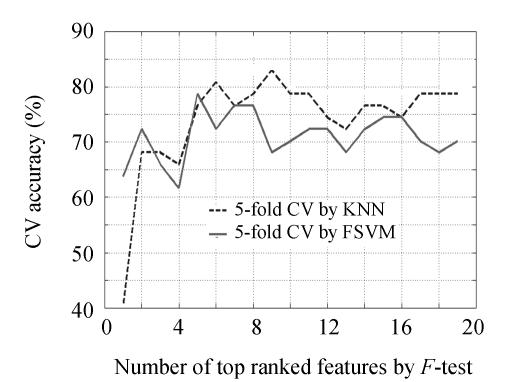
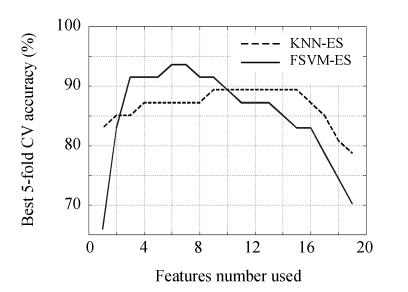
According to the achieved feature rank list, KNN/FSVM-based 5-fold CV accuracy on feature subsets with specific several top features is plotted in Fig.3. Using 5-fold CV by FSVM, optimal feature subset was detected in 5 features (5, 6, 12, 14 and 15) and 78.72% accuracy; using 5-fold CV by KNN, best feature subset was detected in 9 features (5, 6, 7, 8, 11, 12, 14, 15 and 19) and 82.98% accuracy. By F-test method, it may conclude that no distinct feature subset was discovered, and that the optimal feature subset selected by KNN/FSVM-based 5-fold CV performed no better than remarkable single metabolite feature detected.
Fig. 3.
Accuracy curve of KNN/FSVM-based 5-fold CV on specific several top features
Biomarker selection by KNN-ES/FSVM-ES algorithm
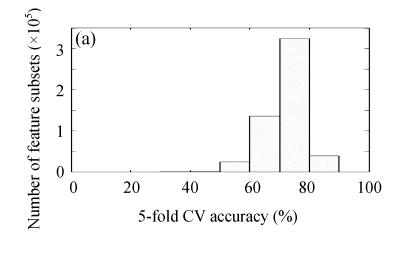
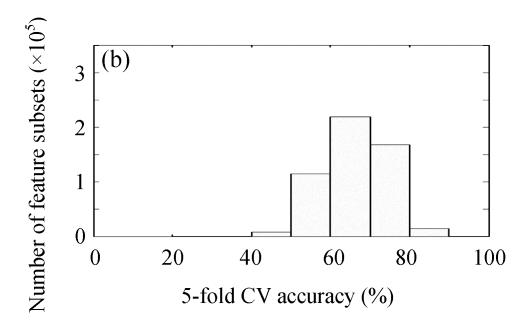
By ES algorithm, 524287 feature subsets were explored. The statistical distribution histograms of the discriminative performance of all feature subsets estimated by KNN/FSVM-based 5-fold CV are plotted in Fig.4 (Fig.4a for KNN-ES and Fig.4b for FSVM-ES). The discriminant performance distribution of feature subsets in KNN-ES is a little more concentrated than that in FSVM-ES. A stricter constraint used in FSVM makes the performance of feature subsets tested in FSVM-ES distributed in wider range. The best discriminant performance for feature subsets with specific feature number found by KNN-ES or FSVM-ES is described in Fig.5. By KNN-ES, the optimal feature subset consisting of 9 features (1, 4, 5, 8, 12, 14, 15, 16 and 17) was found with 89.36% 5-fold CV accuracy. Using KNN combined with optimal feature subset selected by KNN-ES, 2 liver failure samples (1 mild and 1 severe) were misclassified into normal group, and 3 severe liver failure samples were misclassified into mild liver failure group. By FSVM-ES, the optimal feature subset consisting of 6 features (4, 5, 7, 14, 15 and 16) was found with 93.62% 5-fold CV accuracy. Using FSVM combined with optimal feature subset selected by FSVM-ES, no error in classification between normal samples and liver failure samples was found, and only 1 mild liver failure samples was misclassified into severe liver failure group and 2 severe liver failure samples were misclassified into mild liver failure group. The confusion matrices for above two feature subsets are given in Table 2. The optimal feature subsets selected by KNN-ES and FSVM-ES performed far better than that selected by F-test statistics.
Fig. 4.
Statistical distribution histograms of the discriminative performance of all feature subsets in (a) KNN-ES and (b) FSVM-ES
Fig. 5.
Best discriminative performance of feature subsets with specific feature number in KNN-ES and FSVM-ES
Table 2.
Prediction accuracy of optimal feature subsets selected by KNN-ES and FSVM-ES described by confusion matrices
| True classes | Prediction accuracy of optimal feature subsets (%) |
|||||
| KNN-ES |
FSVM-ES |
|||||
| NG | MLFG | SLFG | NG | MLFG | SLFG | |
| NG | 100.00 | 0.00 | 0.00 | 100.00 | 0.00 | 0.00 |
| MLFG | 7.69 | 92.31 | 0.00 | 0.00 | 92.31 | 7.69 |
| SLFG | 7.69 | 27.27 | 63.64 | 0.00 | 18.18 | 81.82 |
NG: Normal group; MLFG: Mild liver failure group; SLFG: Severe liver failure group
By KNN-ES, 291 feature subsets with KNN-based 5-fold CV accuracy greater than 87% were detected in all feature subsets. The sizes of these feature subsets were from 4 to 16. These feature subsets contained 9.9313 features in average, and the standard deviation for the size of these feature subsets was 2.4932. By FSVM-ES, 295 feature subsets with FSVM-based 5-fold CV accuracy greater than or equal to 89% were detected. The sizes of these feature subsets were from 3 to 10. These feature subsets contained 6.6949 features in average, and the standard deviation was 1.4316. The mean discriminant performance for these best feature subsets is described by confusion matrices (Table 3). Best feature subsets selected by FSVM-ES had more balanced error rates among three classes and higher total 5-fold CV accuracy than that done by KNN-ES. Meanwhile, more compact optimal feature subsets were identified by FSVM-ES.
Table 3.
Mean prediction accuracy of several best feature subsets selected by KNN-ES and FSVM-ES described by confusion matrices
| True classes | Mean prediction accuracy of several best feature subsets (%)* |
|||||
| KNN-ES |
FSVM-ES |
|||||
| NG | MLFG | SLFG | NG | MLFG | SLFG | |
| NG | 99.21 | 0.76 | 0.03 | 97.27 | 0.94 | 1.78 |
| MLFG | 7.35 | 92.18 | 0.48 | 5.45 | 89.71 | 4.82 |
| SLFG | 10.72 | 32.40 | 56.89 | 3.61 | 21.91 | 74.48 |
Numbers of the best feature subsets selected by KNN-ES and FSVM-ES are 291 and 295, respectively.
NG: Normal group; MLFG: Mild liver failure group; SLFG: Severe liver failure group
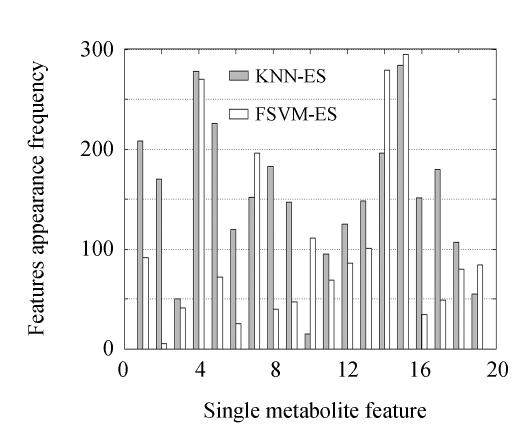
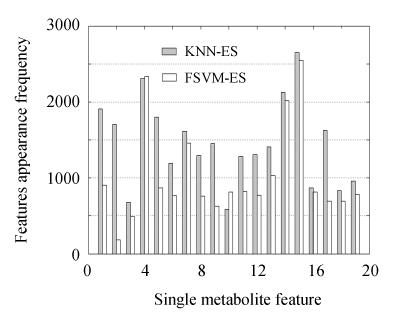
The features’ frequencies in these best feature subsets are plotted in Fig.6. Features 4 and 15 were identified as the most potential biomarkers by KNN-ES and FSVM-ES simultaneously. These two features were also included in the optimal feature subsets selected by KNN-ES and FSVM-ES. All feature subsets with 5-fold CV accuracy greater than or equal to 85% in KNN-ES and FSVM-ES were used as a wider range to analyze the features’ potential as biomarkers. The features’ appearance frequencies in these feature subsets are plotted in Fig.7. Features 4 and 15 were also identified as the two most remarkable features by both of the two algorithms. Feature 14 was another robust discriminative feature found and was also included in the optimal feature subsets selected by KNN-ES and FSVM-ES with a relatively high appearance frequency (Fig.6). By F-test method, Feature 4 was regarded as a noise feature and omitted.
Fig. 6.
Feature appearance frequency in a part of feature subsets with best CV accuracy in KNN-ES and FSVM-ES. There are 291 best feature subsets selected by KNN-ES employed and 295 best feature subsets selected by FSVM-ES employed
Fig. 7.
Feature appearance frequency in feature subsets with 5-fold CV accuracy greater than 85% in KNN-ES and FSVM-ES
Discussion and future work
In this work, the disturbed metabolites were mainly evaluated by the deviation of concentration between the control and diseased patients. Feature 4 corresponds to glyceric acid that is a typical organic acid metabolite in mammalian bio-fluids and exists in two configurations, D(+) and L(−). Both enantiomers are intermediary metabolites normally excreted in trace amount in serum and urine. High level of glyceric acid might be due to some inherited or acquired metabolic disease, especially to the liver and kidney deficiencies. The concentration of glyceric acid in serum samples of the patient group was significantly increased as compared with the healthy group. The higher concentration of glyceric acid may indicate that the metabolisms of the patients were under deterioration and that acidemia might occur in the patients with serious liver failure (Fontaine et al., 1989). Features 14 and 15 correspond to cis-aconitic acid and citric acid, respectively. They are two important intermediary metabolites in tricarboxylic acid (TCA) cycle. The concentration of citric acid in the serum samples of the patient group significantly decreased, while that of cis-aconitic acid increased, as compared with the healthy group. This phenomenon may indicate that the free citric acid in patients’ blood is supplemented into the TCA cycle, and hence augments the concentration of other metabolites in TCA cycle in order to enhance the energy supple (Arai et al., 2001). These results reveal that metabolites are potential as biomarkers for liver failure recognition. Considering that the size of sample dataset is relatively small, the utility of selected biomarkers and their performance should be assessed in a larger size of samples, and more detailed biological and physiological examinations need to be done in the future studies.
In this work, two wrapper algorithms, KNN-ES and FSVM-ES, performed better than the filter algorithm F-test, and FSVM-ES achieved more accurate and compact results than KNN-ES. It may be concluded that the performance of wrapper feature selection method largely depends on the learning algorithm embedded, especially when samples scatter in sparse space, the binary SVM constructed FSVM is more suitable for small dataset, and the optimal feature subset selected by FSVM-ES has stronger discriminative power and more compact construction.
CONCLUSION
In this work, the potential of metabolites as biomarkers for HBV-induced liver failure diagnosis was explored. The serum samples from 24 HBV-induced liver failure patients and 23 healthy volunteers were collected and treated by GC-MS experiments. The 24 patients in different severity were grouped into two classes. Twenty-five commensal peaks in all metabolite profiles were extracted and the relative area values of these peaks were used as features for each sample. Based on the dataset achieved, three feature selection algorithms, F-test, KNN-ES and FSVM-ES, were employed for biomarker identification and seeking optimal feature subset for liver failure diagnosis. On the 3-class dataset with 47 samples and 19 features pre-selected by t-test, 82.98% 5-fold CV accuracy was achieved by 9 features selected by F-test, 89.36% 5-fold CV accuracy by 9 features selected by KNN-ES and 93.62% 5-fold CV accuracy by 6 features selected by FSVM-ES. By analyzing the statistical histogram of features’ appearance frequency in several best feature subsets selected by KNN-ES and FSVM-ES, glyceric acid, cis-aconitic acid and citric acid were proposed as potential diagnostic biomarkers for identifying HBV-induced liver failure. The results indicate that metabolite profile is an effective tool to evaluate the severity of liver diseases, and FSVM-ES is effective for potential diagnostic biomarker selection.
Footnotes
Project supported by the Postdoctoral Science Foundation of China (No. 20070410397), the National Natural Science Foundation of China (No. 60705002) and the Science and Technology Project of Zhejiang Province, China (No. 2005C13026)
References
- 1.Alaoui-Jamali MA, Xu YJ. Proteomic technology for biomarker profiling in cancer: an update. J Zhejiang Univ Sci B. 2006;7(6):411–420. doi: 10.1631/jzus.2006.B0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai K, Lee K, Berthiaume F, Tompkins RG, Yarmush ML. Intrahepatic amino acid and glucose metabolism in a D-galactosamine-induced rat liver failure model. Hepatology. 2001;34(2):360–371. doi: 10.1053/jhep.2001.26515. [DOI] [PubMed] [Google Scholar]
- 3.Dasarathy BV. NN Concepts and Techniques: An Introductory Survey. In: Dasarathy BV, editor. Nearest Neighbour Norms: NN Pattern Classification Techniques. Los Alamitos, CA: IEEE Computer Society Press; 1991. pp. 1–30. [Google Scholar]
- 4.Enas GG, Choi SC. Choice of the smoothing parameter and efficiency of k-nearest neighbor classification. Computers and Mathematics with Applications. 1986;12A(2):235–244. doi: 10.1016/0898-1221(86)90076-3. [DOI] [Google Scholar]
- 5.Fontaine M, Porchet N, Largilliere C, Marrakchi S, Lhermitte M, Aubert JP, Degand P. Biochemical contribution to diagnosis and study of a new case of D-glyceric acidemia/aciduria. Clin Chem. 1989;35(10):2148–2151. [PubMed] [Google Scholar]
- 6.Inza I, Larranaga P, Blanco R, Cerrolaza AJ. Filter versus wrapper gene selection approaches in DNA microarray domains. Artif Intell Med. 2004;31(2):91–103. doi: 10.1016/j.artmed.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM, Kosberg CL, D'Amico G, Dickson ER, Kim WR. A model to predict survival in patients with end-stage liver disease. Hepatology. 2001;33(2):464–470. doi: 10.1053/jhep.2001.22172. [DOI] [PubMed] [Google Scholar]
- 8.Lee WM. Medical progress—hepatitis B virus infection. N Engl J Med. 1997;337(24):1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Zhou M, Han J, Zhu X, Dong T, Gao GF, Tien PJ. Generation of murine CTL by a hepatitis B virus-specific peptide and evaluation of the adjuvant effect of heat shock protein glycoprotein 96 and its terminal fragments. J Immunol. 2005;174:195–204. doi: 10.4049/jimmunol.174.1.195. [DOI] [PubMed] [Google Scholar]
- 10.Luts J, Heerschap A, Suykens JAK, Huffel SV. A combined MRI and MRSI based multiclass system for brain tumour recognition using LS-SVMs with class probabilities and feature selection. Artif Intell Med. 2007;40(2):87–102. doi: 10.1016/j.artmed.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Mao Y, Zhou XB, Pi DY, Wong STC, Sun YX. Multiclass cancer classification by using fuzzy support vector machine and binary decision tree with gene selection. J Biomed Biotechnol. 2005;2005(2):160–171. doi: 10.1155/JBB.2005.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Y, Xia Z, Yin Z, Sun YX, Wan Z. Fault diagnosis based on fuzzy support vector machine with parameter tuning and feature selection. Chin J Chem Eng. 2007;15(2):233–239. doi: 10.1016/S1004-9541(07)60064-0. [DOI] [Google Scholar]
- 13.McConnell JR, Antonson DL, Ong CS, Chu WK, Fox IJ, Heffron TG, Langnas AN, Shaw BW. Proton spectroscopy of brain glutamine in acute liver failure. Hepatology. 1995;22(1):69–74. doi: 10.1002/hep.1840220111. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson JK, Wilson ID. Understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2(8):668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 16.Parker JR. Rank and response combination from confusion matrix data. Information Fusion. 2001;2(2):113–120. doi: 10.1016/S1566-2535(01)00030-6. [DOI] [Google Scholar]
- 17.Theodoridis S, Koutroumbas K. Pattern Recognition. Amsterdam, the Netherlands: Elsevier; 2003. [Google Scholar]
- 18.Vapnik VN. The Nature of Statistical Learning Theory. 2nd Ed. New York, USA: Springer-Verlag; 1999. [Google Scholar]
- 19.Yang J, Xu G, Zheng Y, Kong H, Pang T, Lv S, Yang Q. Diagnosis of liver cancer using HPLC-based metabonomics avoiding false-positive result from hepatitis and hepatocirrhosis diseases. J Chromatogr B. 2004;813(1-2):59–65. doi: 10.1016/j.jchromb.2004.09.032. [DOI] [PubMed] [Google Scholar]