Figure 3. Amino acids of NAP1 required for NAP1 to be glycylated.
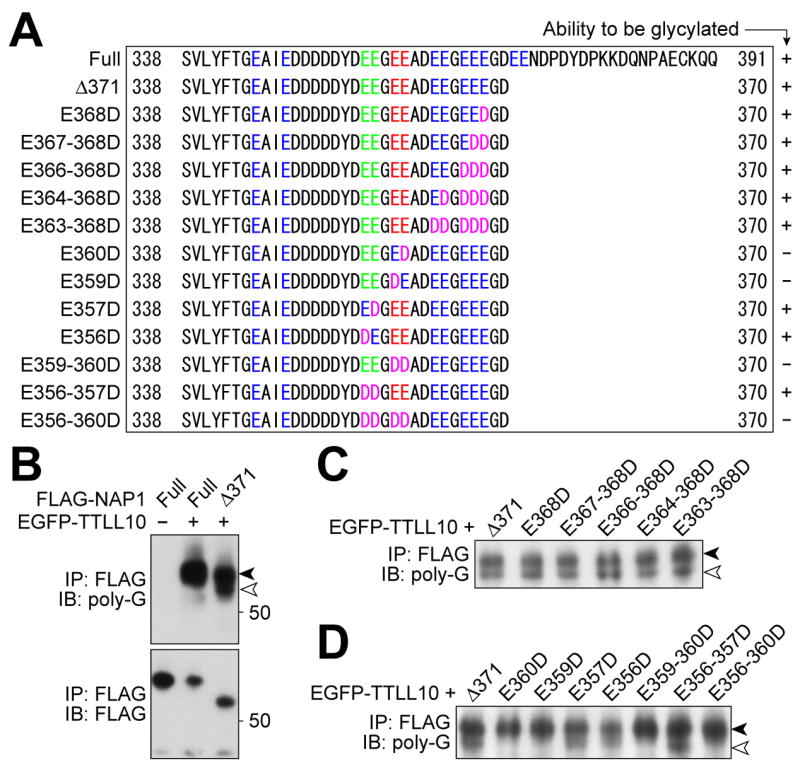
(A) Alignment of C-termini of mouse NAP1 mutants used. Glu (“E”) residues in C-terminal acidic domain are colored. Green text indicates the sites for glutamylation (Regnard et al, 2000). Glu residues colored red were required for glycylation. Asp (“D”) residues replacing Glu are colored magenta. A summary of the relative ability of each protein to be glycylated is shown on the right side. (B-D) Western blot analyses of NAP1 mutants with the R-polygly antibody. The Δ371-NAP1 (white arrowhead) is distinguishable from co-precipitated endogenous NAP1 (black arrowhead), with both species detected by the R-polygly antibody (B). Substitution of all Glu residues between 363 and 368 with Asp did not affect the ability to be a substrate for TTLL10 (C). Replacement of either Glu359 or 360 by Asp is sufficient to cause a loss of reactivity to the R-polygly antibody, while substituting the glutamylation sites with Asp residues did not impair the NAP1 ability to be glycylated by TTLL10 (D).
