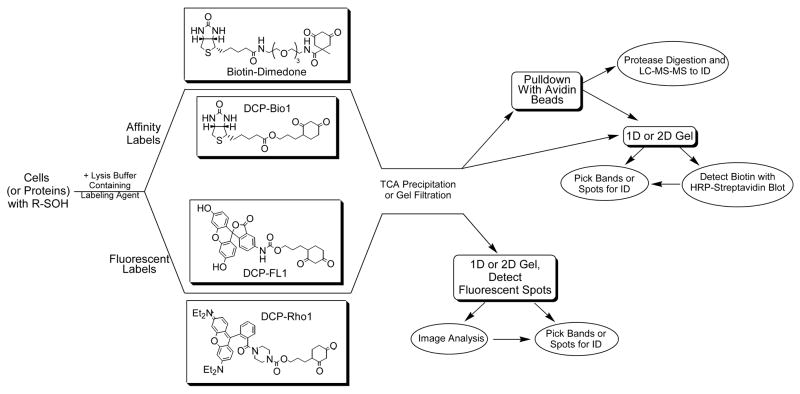
Figure 3. Strategies for detecting and isolating sulfenic acids in proteins.
Cells or proteins with cysteine sulfenic acid modifications are incubated with either affinity (biotin-dimedone or DCP-Bio1, or two other affinity probes [39,41]) or fluorescently-tagged reagents (DCP-FL1 or DCP-Rho1, or two other fluorescein- and rhodamine-based probes [39]). The chemically reactive probes based on dimedone include a nucleophilic carbon between the two carbonyls of the cyclohexane ring that exhibits specificity toward sulfenic acids. After incubation, unreacted probes are removed from the protein samples by trichloroacetic acid (TCA) precipitation or gel filtration chromatography. Subsequent analytical procedures can include one-dimensional or two-dimensional gels in both cases, from which bands or spots can be excised, digested and analyzed by mass spectrometry (MS) for identification of the labeled protein. For the biotinylated samples, “pulldown” of the affinity-labeled proteins with avidin-linked beads can be carried out either before or after proteolytic digestion of the proteins in the samples, enriching in either labeled proteins or labeled peptides. Subsequent LC-MS-MS analysis can then be used to identify labeled proteins/peptides.