Fig. 4. ECM eQTL gene expression signatures predict survival in human breast cancer.
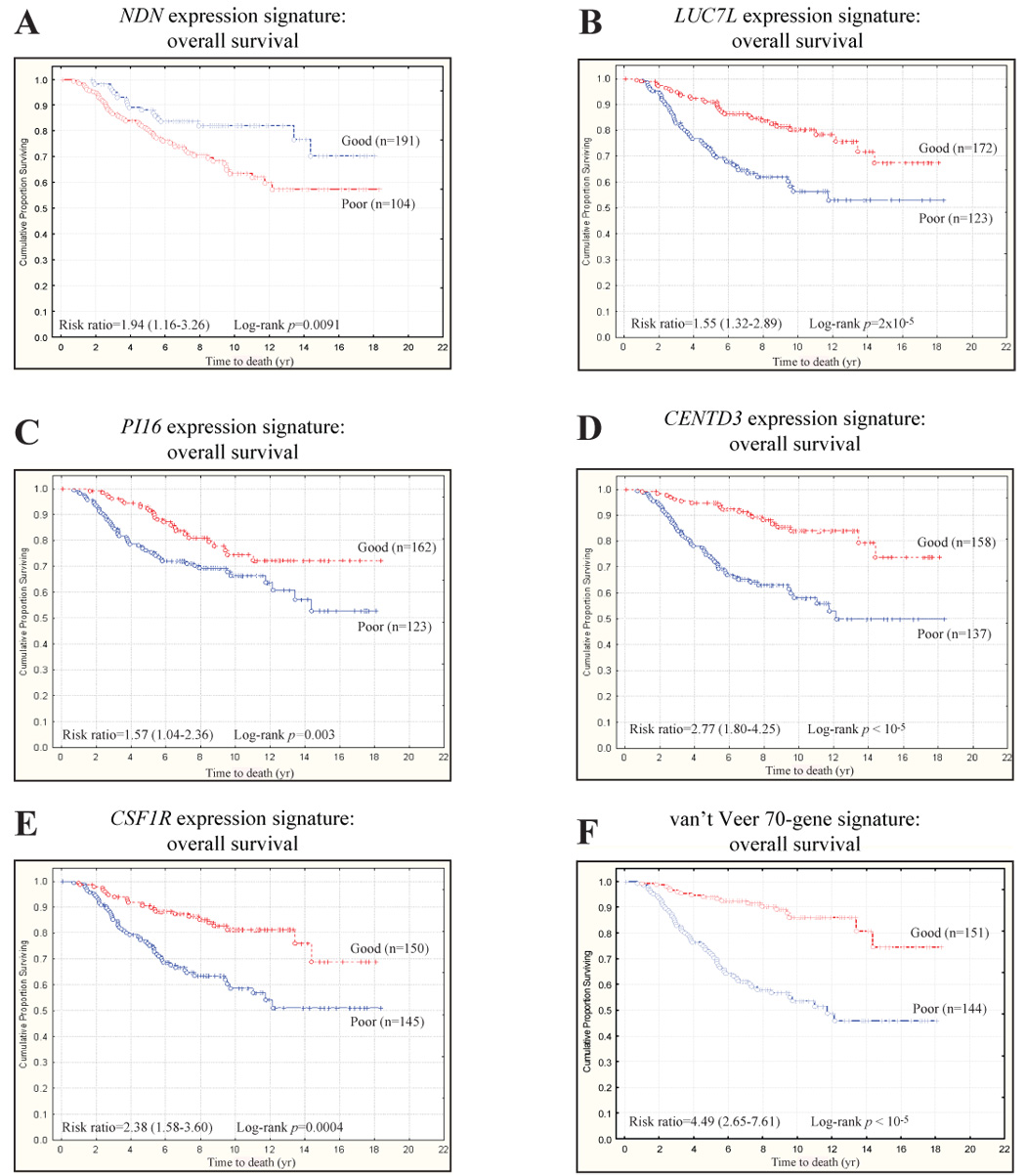
Diasporin Pathway candidate genes accurately predict overall survival in the Dutch Rosetta dataset. (a) The cumulative survival for the NDN signature was estimated to be 70% vs. 57% for the good and poor prognosis NDN signatures, respectively (NDN signature hazard ratio = 1.94, 95% confidence interval [CI] = 1.16–3.26). (b) The cumulative survival for the LUC7L signature was estimated to be 67% vs. 53% for the good and poor prognosis LUC7L signatures, respectively (LUC7L signature hazard ratio = 1.55, 95% CI = 1.32–2.89). (c) The cumulative survival for the PI16 signature was estimated to be 72% vs. 53% for the good and poor prognosis PI16 signatures, respectively (PI16 signature hazard ratio = 1.57, 95% CI = 1.04–2.36). (d) The cumulative survival for the CENTD3 signature was estimated to be 74% vs. 50% for the good and poor prognosis CENTD3 signatures, respectively (CENTD3 signature hazard ratio = 2.77, 95% CI = 1.80–4.25). (e) The cumulative survival for the CSF1R signature was estimated to be 69% vs. 51% for the good and poor prognosis CSF1R signatures, respectively (CSF1R signature hazard ratio = 2.38, 95% CI = 1.58–3.60). (f) Indeed, it appears that these candidate gene signatures have a similar ability to predict survival in this dataset than the 70-gene signature described by van’t Veer et al [4]. Specifically, the survival for the good and poor prognosis 70-gene signatures was estimated to be 73% vs. 47%, respectively (70 gene signature hazard ratio = 4.49, 95% CI = 2.65–7.61).
