Figure 4.
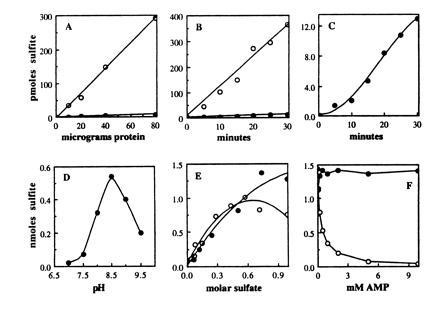
Kinetics of sulfite formation by the APR1 enzyme. (A–C) The open circles reflect reactions with APS as a substrate and the solid circles reflect reactions with PAPS as a substrate. All the abscissas are labeled as in A. (A) Sulfite formed in 15 min at 30°C by varying amounts of protein from an extract of JM96 carrying APR1. (B) Sulfite formation over time in reactions containing 40 μg of protein from an extract of JM96 carrying APR1. (C) The data of graph in C with the scale of the abscissa expanded 30-fold. The sulfonucleotide reductase reaction conditions were as described in Table 2. (D–F) Optimization of APR1-mediated APS reductase activity. All the abscissas are labeled as in D. (D) pH optimum. The reaction mixture contained in a volume of 100 μl: 100 mM Tris·HCl, 500 mM Na2SO4, 1 mM EDTA, 5 mM DTT, 25 μM [35S]APS (≈500 Bq × nmol−1) and 20 μg (2 μl) cell extract. The reaction was incubated for 20 min at 30°C. The pH was adjusted in a 2× mixture of Tris, Na2SO4, and EDTA; then water, DTT, APS, and extract were sequentially added to initiate the reaction. Protein extract was prepared in 50 mM Tris·HCl (pH 8.0) from E. coli strain A522 expressing APR1. (E) Optimal salt concentration. The reaction mixture was as described in D, but with 50 mM Tris·HCl (pH 8.5), 40 μg (4 μl) cell extract from E. coli strain JM96 expressing APR1, and the indicated concentration of Na2SO4 (solid circles) or (NH4)2SO4 (open circles). (F) Inhibition by 5′-AMP. The reaction was as described in E, but with 40 μg (4 μl) cell extract from E. coli strain A522 expressing APR1, and 5′-AMP (open circles) or 2′-AMP (solid circles) at the indicated concentration.
