Abstract
We used biotinylation and streptavidin affinity chromatography to label and enrich proteins from apical and basolateral membranes of rat kidney inner medullary collecting ducts (IMCDs) prior to LC-MS/MS protein identification. To enrich apical membrane proteins and bound peripheral membrane proteins, IMCDs were perfusion-labeled with primary amine-reactive biotinylation reagents at 2 °C using a double barreled pipette. The perfusion-biotinylated proteins and proteins bound to them were isolated with CaptAvidin-agarose beads, separated with SDS-PAGE, and sliced into continuous gel pieces for LC-MS/MS protein identification (LTQ, Thermo Electron Corp.). 17 integral and glycosylphosphatidylinositol (GPI)-linked membrane proteins and 44 non-integral membrane proteins were identified. Immunofluorescence confocal microscopy confirmed ACVRL1, H+/K+-ATPase α1, NHE2, and TauT expression in the IMCDs. Basement membrane and basolateral membrane proteins were biotinylated via incubation of IMCD suspensions with biotinylation reagents on ice. 23 integral and GPI-linked membrane proteins and 134 non-integral membrane proteins were identified. Analyses of non-integral membrane proteins preferentially identified in the perfusion-biotinylated and not in the incubation-biotinylated IMCDs revealed protein kinases, scaffold proteins, SNARE proteins, motor proteins, small GTP-binding proteins, and related proteins that may be involved in vasopressin-stimulated AQP2, UT-A1, and ENaC regulation. A World Wide Web-accessible database was constructed of 222 membrane proteins (integral and GPI-linked) from this study and prior studies.
The renal collecting duct is the terminal part of the renal tubule. Its major function is to transport water and solutes in a regulated manner. Although there are many regulatory factors that affect collecting duct transport functions, one of the most important factors is vasopressin, a peptide hormone secreted by the posterior pituitary gland. Vasopressin regulates several transport proteins including aquaporin 2 (1, 2), aquaporin 3 (3), the epithelial sodium channel ENaC1 (4, 5), and the urea transporter UT-A (6, 7). Abnormalities of regulatory processes in the collecting ducts are responsible for a large number of clinically important disorders of salt and water balance (8, 9).
The terminal portion of the collecting duct is the IMCD. Investigation of the mechanisms of vasopressin action in the IMCD is benefiting from analysis of the proteome of the IMCD cells. The work so far has identified a large number of IMCD proteins that have been included in a publicly accessible database, the IMCD Proteome Database (dir.nhlbi.nih.gov/papers/lkem/imcd/index.htm). Because most of the proteomics methods used so far (10–13) to construct this database are biased against integral membrane and glycosylphosphatidylinositol (GPI)-linked membrane proteins, these classes of proteins appear underrepresented. The difficulty in detecting membrane proteins has arisen largely because of the difficulty of solubilizing them in detergents that are compatible with two-dimensional electrophoresis. However, shotgun proteomics using LC-MS/MS offers greater efficiency in identification of integral membrane proteins because they can be solubilized using the strong ionic detergent SDS and then separated on one-dimensional gels prior to trypsinization. However, biochemical approaches are needed to isolate specific membrane fractions prior to identification. One objective of the present study was to devise an approach that increases identification of integral membrane proteins and GPI-linked proteins in plasma membrane domains as well as proteins that are bound to integral membrane proteins.
Plasma membrane segregation into apical and basolateral domains at the tight junctions provides the key functionality of epithelial cells. Discrete proteomes are expected in the apical and the basolateral membranes to account for structural and functional differences including hormone responses and vectorial transport across the epithelia. In the kidney, Cutillas et al. (14) were the first to profile the apical and basolateral proteomes of renal cortex tissue using samples prepared from differential centrifugation and free flow electrophoresis. Because of the relative abundance of proximal tubules in the renal cortex, the findings from this study are probably applicable to the proximal tubules but not the collecting ducts. Here we devised methods combining surface biotinylation and streptavidin affinity chromatography to label and enrich proteins from apical and basolateral membranes of IMCDs prior to LC-MS/MS protein identification. 62 integral and GPI-linked membrane proteins were identified. Subtractive comparison of non-integral membrane proteins identified in the apical and not the basolateral membrane revealed 25 potential signaling and trafficking proteins involved in vasopressin-regulated AQP2, UT-A, and ENaC regulation.
EXPERIMENTAL PROCEDURES
Animals
Pathogen-free male Sprague-Dawley rats (Taconic Farms Inc., Germantown, NY) were maintained on ad libitum rat chow (NIH-07; Zeigler, Gardners, PA) and drinking water in the Small Animal Facility, NHLBI, National Institutes of Health. Animal experiments were conducted under the auspices of the animal protocol H-0110 approved by the Animal Care and Use Committee, NHLBI, National Institutes of Health. Adult animals weighing between 200 and 250 g were injected intraperitoneally with furosemide (5 mg/rat) 20 min before decapitation and removal of kidneys. Furosemide dissipates the medullary osmolality, thereby preventing osmotic shock to the cells upon isolation of the inner medullae (15). Immediately after the inner medullae were excised from the kidneys, they were transferred in ice-cold isolation solution (250 mM sucrose, 10 mM Tris, pH 7.4) to a cold room (2 °C) for apical surface biotinylation. Some excised inner medullae were used to prepare IMCD suspensions for basolateral surface biotinylation.
Perfusion Biotinylation of IMCDs
In the cold room, each inner medulla was placed on a porous support that allows drainage of excess fluid and in between two stacks of filter papers that moisturize the tissue (Fig. 1A). To introduce biotinylation reagents to the lumens of IMCDs, a double barreled pipette was made from a theta glass capillary (TST 150-6; World Precision Instruments, Inc., Sarasota, FL). The tip of the pipette was bent close to 90° and drawn to a spindle shape with a diameter of 100 μm tapering to 30 μm at its opening to fit the openings of the IMCDs (ducts of Bellini) at the inner medullary tip. The geometry of the pipette tip was made such that the body of the spindle seals the duct of Bellini to prevent backflow of the perfused fluid. Two surface biotinylation reagents (thiol-cleavable sulfo-NHS-SS-biotin or non-cleavable sulfo-NHS-LC-biotin, Pierce) were used in different experiments at a concentration of 1.5 mg/ml in PBS (5.1 mM Na2HPO4, 1.2 mM KH2PO4, 154 mM NaCl, pH 7.4). These reagents covalently link biotin to surface proteins through the N-hydroxysuccinimide (NHS) group that reacts with primary amines at the N′ termini and on side chains of lysine residues. They are believed to be excluded from the cell interior due to their negatively charged sulfonate groups. However, our preliminary experiments showed that two of these nominally “membrane-impermeant” reagents readily entered the IMCD cells. Preliminary experiments also showed that a brief fixation of the cell membrane lipids with 4% paraformaldehyde (16) prior to biotinylation prevented intracellular biotinylation at 2 °C (Fig. 1, C and D). The optimized labeling process took place in the following sequence. One barrel delivered the fixative to the IMCD lumen for 5 min followed by the other barrel delivering the biotinylation reagent (sulfo-NHS-LC-biotin) for another 5 min. After the biotinylation step, the solution was switched back to the fixative that flushed out the biotinylation reagents and stayed in the lumen while other ducts of Bellini were being perfusion-biotinylated. On average, between six and eight ducts of Bellini of a single inner medulla were perfusion-biotinylated. Two non-toxic food dyes (FD&C Blue No. 1 and FD&C Red No. 3) were used in the perfusates to visualize solution change. Another pipette (single barreled) was situated at the top of the perfusion pipette to drip Tris-buffered isolation solution onto the tissue to moisturize the tissue and to quench the reactive NHS group of the biotinylation reagents if backflow occurred. After perfusion-biotinylation, the inner medullae were immediately frozen on dry ice. A total of 12 inner medullae from six rats were collected. Some non-fixed inner medullae were perfused with sulfo-NHS-SS-biotin. Both fixed and non-fixed perfusion-biotinylated IMCDs were prepared for LC-MS/MS analysis in different experiments.
Fig. 1. Apical surface biotinylation via retrograde perfusion of IMCDs.
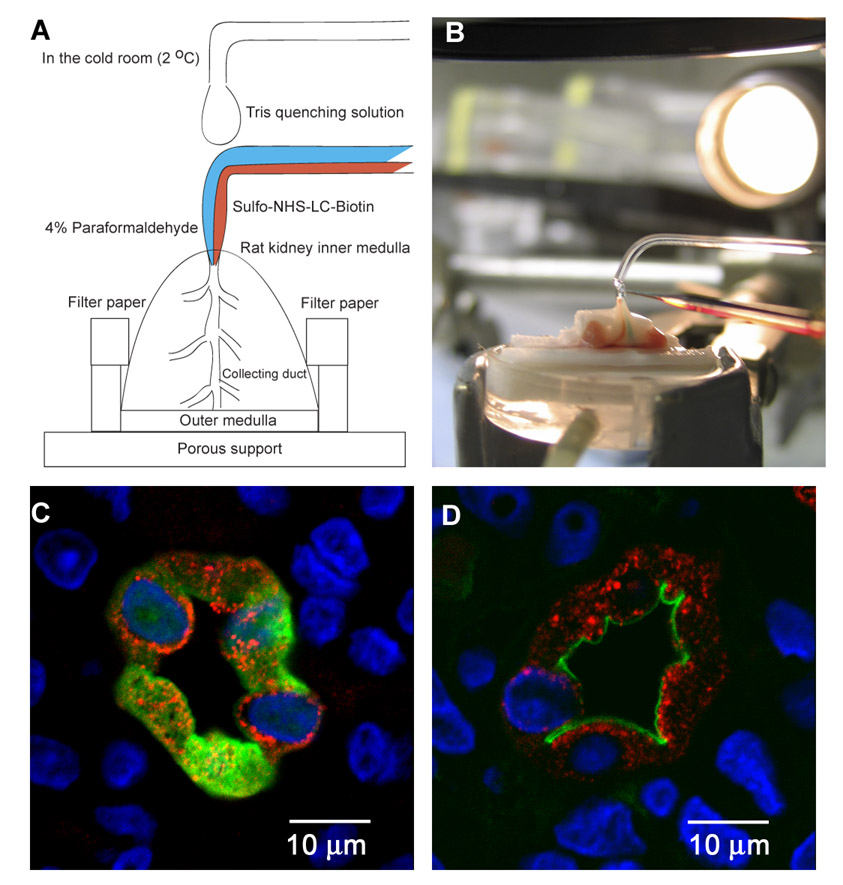
A, a rat kidney medulla was excised and placed on a porous support that allows fluid drainage and in between filter papers that moisturize the tissue. Apical membrane proteins were labeled with biotin via perfusing the IMCD lumens using a custom-made double barreled pipette in a cold room (2 °C) to inhibit endocytosis. One barrel delivered paraformal-dehyde to fix the membrane lipids before the other barrel delivered sulfo-NHS-LC-biotin or sulfo-NHS-SS-biotin to label the apical membrane proteins. The fixation of lipids prior to biotinylation of proteins was necessary to ensure apical protein biotinylation. Blue and red food dyes were used in the perfusates to visualize fluid change in the IMCDs. A single barreled pipette at the top of the perfusion pipette supplied Tris buffer to moisturize the tissue and to quench the biotinylation reagent if backflow occurred. B, a picture shows the perfusion setup in the cold room. C, a fluorescence micrograph shows biotinylation occurring throughout the perfused IMCD cells without prior fixation revealed by streptavidin-FITC that stains biotin in green. The IMCD marker AQP2 staining and the DAPI nuclear staining are shown in red and blue, respectively. D, a fluorescence micrograph shows restricted apical membrane biotinylation in the perfused IMCDs with prior fixation.
Basolateral Biotinylation of IMCDs
To label the basement membrane and basolateral membrane proteins with biotin, an IMCD suspension was prepared from the excised inner medullae as described previously (17). Briefly the inner medullae were minced and digested with 2 mg/ml hyaluronidase and 3 mg/ml collagenase B. A 60 × g centrifugation was then carried out to precipitate the heavier IMCD segments from the non-IMCD components of the inner medulla (loops of Henle, interstitial cells, vasa recta, and capillaries). The isolated IMCD suspension was fixed with 4% paraformaldehyde for 5 min on ice before incubation with 1.5 mg/ml sulfo-NHS-LC-biotin for 5 min on ice to label selectively the basement membrane and basolateral membrane proteins as described previously (18). Another IMCD suspension that was not fixed was labeled with 1.5 mg/ml sulfo-NHS-SS-biotin on ice for 5 min. Both fixed and non-fixed incubation-biotinylated IMCD suspensions were used for LC-MS/MS protein identification in different experiments.
Plasma Membrane-enriched Fraction
To enrich for plasma membrane components of the IMCD cells, a high density membrane fraction was prepared using differential centrifugation as described previously (3, 19). Perfusion-biotinylated inner medullae were homogenized in liquid nitrogen using a mortar and a pestle. The inner medulla homogenate was suspended in ice-cold isolation buffer containing protease inhibitors (0.1 mg/ml PMSF and 1 μg/ml leupeptin) and centrifuged at 1,000 × g for 10 min at 4 °C to remove incompletely homogenized fragments and nuclei. The supernatant was collected and centrifuged again at 17,000 × g for 20 min. The 17,000 × g pellet is a high density membrane fraction that was reported previously to be enriched for plasma membrane (19).
Incubation-biotinylated IMCD suspensions were homogenized in the ice-cold isolation buffer containing the protease inhibitors using a tissue homogenizer (TH; Omni International, Marietta, GA). The high density membrane fraction was prepared as described above.
Isolation of Biotinylated Proteins
When the non-cleavable biotinylation reagent was used, high density membrane fractions were prepared, and the membranes were solubilized with 1 ml of lysis solution (150 mM NaCl, 5 mM EDTA, 50 mM Tris, pH 7.4) containing 1% Nonidet P-40 plus protease inhibitors (0.1 mg/ml PMSF and 1 μg/ml leupeptin). The solubilized membrane fraction was centrifuged at 10,000 × g for 10 min at 4 °C to remove insoluble components. 100 μl of the resulting supernatant was saved as preisolation control, and 900 μl of it was mixed with 200 μl of sediment CaptAvidin-agarose beads (Invitrogen) that bind biotinylated proteins (20). After removal of unbound proteins, the CaptAvidin-agarose beads were washed with the following solutions (900 μl for each wash) to remove nonspecifically bound proteins: 1) lysis solution containing 1% Nonidet P-40 three times, 2) high salt solution (500 mM NaCl, 5 mM EDTA, 50 mM Tris, pH 7.4) two times, and 3) no salt solution (10 mM Tris, pH 7.4) one time. The biotinylated proteins were eluted from the CaptAvidin-agarose beads twice, each time with 45 μl of alkaline solution (50 mM Na2CO3, pH 10.1) plus 10 mM D-biotin (Invitrogen) that competes the biotin binding sites on the CaptAvidin molecules thereby enhancing elution of biotinylated proteins (20).
When the thiol-cleavable biotinylation reagent was used, the isolation procedures for the biotinylated proteins were similar to those for the non-cleavable biotinylated proteins except 1) radioimmune precipitation assay detergent solution (0.1% SDS, 0.5% sodium de-oxycholate, and 1% Nonidet P-40) was used in the lysis buffer and the washing buffer, 2) streptavidin-agarose beads (Pierce) were used, and 3) the elution was done via cleaving the disulfide bond with lysis buffer containing 100 mM DTT for 30 min at room temperature.
Preparation of Proteins for Mass Spectrometric Identification
The purified biotinylated proteins were concentrated with 10,000-Da cutoff Microcon centrifugal filter devices (YM-10; Millipore Corp., Bed-ford, MA) and separated by SDS-PAGE using 10% polyacrylamide minigels (Bio-Rad) to reduce sample complexity. Gels were silver-stained using SilverQuest™ (Invitrogen) or GelCode (Pierce) to visualize the proteins. The entire sample lane was cut into 15–16 sequential slices of about 2-mm thickness. Proteins in each gel slice were destained, reduced, alkylated, and trypsin-digested using a protocol described previously (21).
LC-MS/MS Protein Identification
Tryptic peptides extracted from each gel slice were injected using an Agilent 1100 nanoflow system (Agilent Technologies, Palo Alto, CA) into a reversed-phase liquid chromatographic column (PicoFrit™, Biobasic C18; New Objective, Woodburn, MA) to further reduce sample complexity before mass analyses using an LTQ mass spectrometer (Thermo Electron Corp., San Jose, CA) equipped with a nanoelectrospray ion source. The m/z ratios of peptides and their fragmented ions were recorded as spectra by the mass spectrometer. The spectra with a total ion current greater than 10,000 were used to search for matches to peptides from rat proteins (4,296 entries) in the Swiss-Prot database using the Bio-works software (Version 3.1; Thermo Electron Corp.) based on the Sequest algorithm. The search parameters included: 1) precursor ion mass tolerance less than 2 amu, 2) fragment ion mass tolerance less than 1 amu, 3) up to three missed tryptic cleavages allowed, and 4) amino acid modifications cysteine carboxyamidomethylation (plus 57.05 amu), methionine oxidation (plus 15.99 amu), and lysine biotinylation (plus 89.00 amu for residual thiol-cleavable biotin mass or plus 339.00 amu for non-cleavable biotin mass). Matched peptide sequences must pass the following filters for provisional identification: 1) the cross-correlation scores (Xcorr) of matches were greater than 1.5, 2.0, and 2.5 for charged state 1, 2, and 3 peptide ions, respectively, 2) the uniqueness scores of matches (ΔCn) were higher than 0.08, and 3) the ranks of the primary scores (Rsp) were less than 10. To assess the overall quality of peptide matches, the same spectra were used to search for random matches using the same searching parameters, filter settings, and database except the sequences in the database were reversed. The random match results served as references for accepting spectra after manual examinations. For those peptides that passed the filters, the spectra with the highest Xcorr were manually examined to confirm protein identification. 38 examples of the accepted spectra are listed in Supplemental Table S-1. Most of these spectra are selected from the fixed perfusion-biotinylated IMCDs (n = 30). Regardless of this manual spectrum inspection, the only proteins that are presented under “Results” are those with high quality spectra for two or more distinct peptide ions or single-peptide identifications that were confirmed with either immunofluorescence staining or RT-PCR. However, all single-peptide identifications that passed manual inspection are presented in Supplemental Tables S-2, S-4, and S-6.
Immunofluorescence Confocal Microscopy
Adult rats were treated with furosemide before decapitation and removal of the kidneys as described above. Kidney slices containing cortex and medulla were fixed with 4% paraformaldehyde overnight and processed using an automated tissue-embedding console system (Tissue-Tek VIP™ 5 and Tissue-Tek TEC™ 5; Sakura Finetek, Torrance, CA) using paraffin as the embedding agent. Tissue sections (4 μm) were obtained using a microtome (RM2125; Leica Microsystems Inc., Bannockburn, IL). The sections were rehydrated, and antigen retrieval was carried out with microwave heat for 15 min in TEG buffer (10 mM Tris and 0.5 mM EGTA, pH 9.0). After neutralization with NH4Cl buffer, the sections were blocked with 1% BSA, 0.2% gelatin, and 0.05% saponin in PBS before incubation overnight with primary antibody diluted in 0.1% BSA and 0.3% Triton X-100 in PBS. The rabbit and chicken primary antibody against AQP2 (LL127 and LL265) were generated in our laboratory (1, 22). Other primary antibodies were gifts or commercial products: H+/K+-ATPase α1 (Adam Smolka, Medical University of South Carolina, Charleston, SC), TauT (Russell Chesney, University of Tennessee Health Science Center, Memphis, TN), ACVRL1 (Abgent Inc., San Diego, CA), and NHE2 (Alpha Diagnostics International Inc., San Antonio, TX). After rinsing with 0.1% BSA, 0.2% gelatin, and 0.05% saponin in PBS, the sections were incubated for 1 h with secondary antibody diluted in 0.1% BSA and 0.3% Triton X-100 in PBS. The secondary antibodies used were FITC-, Alexa488-, or Alexa568-conjugated (Invitrogen). After washing with PBS, the sections were mounted in Vectashield solution containing DAPI to stain nuclei (H-1500; Vector Laboratories, Burlingame, CA). Confocal fluorescence images were taken using a Zeiss LSM 510 microscope and software (Carl Zeiss MicroImaging, Inc., Thornwood, NY). Immunostaining without primary antibody served as a negative control and yielded no staining.
Sites of biotinylation in the perfusion- or incubation-biotinylated IMCDs were examined with streptavidin-FITC staining and confocal microscopy. Perfusion-biotinylated inner medullae were fixed with 4% paraformaldehyde, processed, embedded, and sectioned as described above. After rehydration, the sections were stained with streptavidin-FITC for 1 h, washed with PBS twice, and mounted in the Vectashield medium for observation. Incubation-biotinylated IMCDs were fixed with 4% paraformaldehyde for 10 min. After removal of the fixative, the IMCDs were suspended in 50 μl of OCT compound (Sakura Finetek) and frozen at the bottom of the Cryomold (Sakura Finetek). Additional OCT compound was gradually added to the frozen IMCD suspension to make the IMCD suspension block. 4-μm sections were obtained from the block using a Leica CM3050 S cryostat. The sections were fixed with −20 °C methanol for 5 min, dried, washed with PBS twice for 5 min each, and stained with streptavidin-FITC for 1 h. After a wash with PBS, the sections were mounted in the Vectashield medium for observation.
RT-PCR
IMCD suspensions were prepared as described above. Total RNA of the IMCD suspensions was extracted using TRIzol® reagent (Invitrogen). First strand cDNA was produced from the total RNA extract (5 μg) using an oligo(dT) primer and SuperScript™ reagents (Invitrogen) in a reaction volume of 20 μl. PCR product was generated from 0.5 μl of the first strand cDNA mixture using gene-specific primers (Supplemental Table S-3) and the ImmoMix (Bioline USA Inc., Randolph, MA). The PCR was performed on a Peltier PCT-200 thermal cycler (MJ Research, Waltham, MA) with the following conditions: activation of the DNA polymerase at 95 °C for 7 min; 30 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s; and a final extension at 72 °C for 10 min. The RT-PCR products were resolved with 1.6% agarose gel electrophoresis, stained with ethidium bromide, and visualized using the Gel Logic 100 imaging system (Eastman Kodak Co.).
RESULTS
Entry of Biotinylation Reagents into IMCD Cells
The strategy used in this study was to use surface biotinylation and streptavidin chromatography to label and enrich proteins in the apical or basolateral plasma membrane and then to identify them using combined LC-MS/MS. The strategy was designed to identify integral membrane proteins and GPI-linked proteins as well as peripheral membrane proteins bound to the labeled proteins. To enrich proteins of the apical plasma membrane, we perfused rat IMCDs from the ducts of Bellini at the tip of isolated renal medullae with biotinylation reagents (Fig. 1, A and B). Perfusion from a single duct of Bellini resulted in labeling of a large number of IMCDs because of the merging organization of the collecting ducts in the inner medulla. Although the biotinylation reagent sulfo-NHS-SS-biotin chosen for this task is classified as a membrane-impermeant form and the tissue processing and perfusion was done at 2 °C, staining with streptavidin-FITC revealed that the reagent entered the IMCD cells, resulting in diffuse biotinylation distributed throughout the cells (Fig. 1C, green). Another similar biotinylation reagent sulfo-NHS-LC-biotin yielded the same results (data not shown), indicating a common problem with these reagents. The biotinylation, however, was restricted to the perfused IMCD cells (compare with distribution of a collecting duct cell protein AQP2, red).
As shown in Fig. 1D, when the IMCDs were first perfused with a fixative (4% paraformaldehyde) to fix the lipid and protein components of the cells (16), the biotinylation was limited to the apical plasma membrane. Thus, the technique can be used to label all proteins in the IMCD cells if used without prior fixation or to selectively label apical plasma membrane proteins if used with prior fixation. Because the non-ionic detergents used in the affinity isolation of biotinylated proteins do not in general disrupt protein-protein binding interactions, this technique allows isolation of both biotinylated proteins and other proteins that are bound in complexes with the biotinylated proteins.
Proteomics Analyses of Fixed Perfusion-biotinylated IMCDs
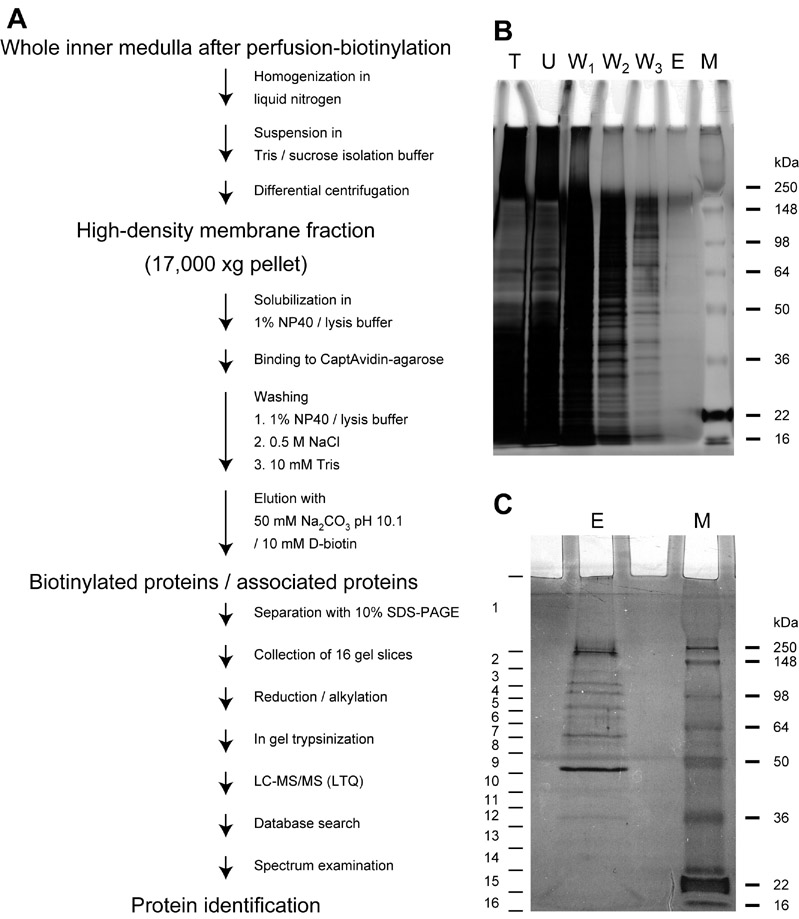
Fig. 2A shows the purification process of biotinylated proteins from the fixed perfusion-biotinylated IMCDs. In general, we perfused between six and eight ducts of Bellini in each inner medulla of a total between 10 and 12. Fig. 2B shows a silver-stained gel image of the proteins prepared from the fixed perfusion-biotinylated IMCDs, indicating that the final eluate (lane E) from the CaptAvidin-agarose beads contained but a very small fraction of the total protein present in the original sample (lane T). Fig. 2C shows a silver-stained gel image of the concentrated eluate (lane E), indicating how the gel was cut into 16 slices (slice numbers are on the left) prior to in-gel trypsinization and protein identification using LC-MS/MS.
Fig. 2.
A, a flow chart summarizing the isolation, preparation, and identification of proteins in the fixed perfusion-biotinylated IMCDs. B, a silver-stained image showing the isolation processes of biotinylated proteins and their associated proteins in the fixed perfusion-biotinylated IMCDs. The total membrane fraction (lane T) was prepared and bound to the CaptAvidin-agarose beads. After removal of the unbound (U) and the non-specifically bound proteins in three washing buffers (W1–3), the biotinylated proteins and their associated proteins were eluted (E) from the CaptAvidin-agarose beads. C, a silver-stained image showing how the biotinylated proteins and their associated proteins were separated with SDS-PAGE and sliced into 16 pieces (numbers on the left) to reduce sample complexity prior to preparation for LC-MS/MS protein identification. Lane M, molecular mass markers.
Table I summarizes the number of proteins identified by LC-MS/MS from all slices added together. The first column of Table I indicates the number of peptides that were identified for particular proteins. The second column of Table I (labeled “Original database search”) indicates the number of proteins identified by a standard computer search using the Swiss-Prot database and Bioworks software with default settings (see “Experimental Procedures”). As shown, most identifications with these settings would be based on matches of a single tryptic peptide. An important objective in protein mass spectrometry is to devise approaches that eliminate or minimize false positive identifications. Toward this end, we carried out computational control experiments in which the same spectra were analyzed in exactly the same way except that the database elements consisted of reversed sequences from the Swiss-Prot database. The number of proteins putatively identified in this control experiment is summarized in the third column of Table I labeled “Reversed database search.” As can be seen, there was a high rate of putative “identifications” derived from randomly matched sequences. The fourth column of Table I labeled “Net difference” shows the numbers in the second column subtracted from the numbers in the third column. The comparison indicates that, for identifications based on one- or two-peptide matches, a relatively small fraction is likely to be valid based only on computerized identification. Therefore, to efficiently eliminate false positive identifications, we manually examined all spectra to be included in the results reported in this study (last column labeled “Manually accepted identification”). Criteria for the acceptance are described under “Experimental Procedures.” Because many of these potential identifications were single-peptide identifications, we required positive results for either immunofluorescence staining or RT-PCR to confirm the expression of these proteins (Supplemental Table S-1). However, for completeness, all manually inspected single-peptide identifications are reported in Supplemental Table S-2.
Table I.
Number of proteins identified from the fixed perfusion-biotinylated inner medullary collecting ducts
| Peptide numbera | Original database searchb | Reversed database searchc | Net differenced | Manually accepted identificatione |
|---|---|---|---|---|
| 1 | 378 | 263 | 115 | 75 |
| 2 | 39 | 24 | 15 | 21 |
| 3 | 12 | 3 | 9 | 8 |
| 4 | 5 | 1 | 4 | 4 |
| >4 | 4 | 0 | 4 | 3 |
| Total | 438 | 291 | 147 | 111 |
Number of peptides identified for particular proteins.
Number of proteins passing the default criteria of the Bioworks/Sequest search algorithm using the Swiss-Prot database.
Same as Footnote b except the sequences in the database were reversed.
Estimated number of proteins likely to be valid identifications.
Number of proteins passing manual spectrum inspection.
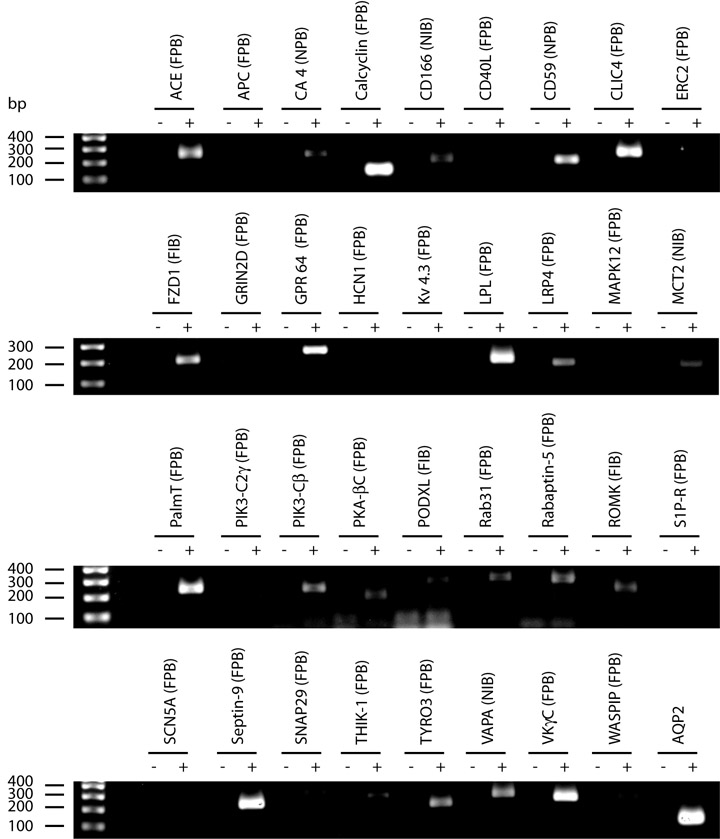
Fig. 3 shows immunofluorescence localization in the renal inner medulla of four of the proteins identified in the fixed perfusion-biotinylated IMCDs for which antibodies could be obtained, viz. ACVRL1, H+/K+-ATPase α1, NHE2, and TauT. ACVRL1 was identified with two peptides, and the other three proteins were identified with one peptide. Each was found to co-localize with AQP2, a collecting duct marker protein, confirming their presence in the IMCD. For protein identifications that lack available antibodies, RT-PCR was used to examine their mRNA expression in the isolated IMCD suspensions. As shown in Fig. 4, 27 primer pairs designed for the single-peptide identifications from the fixed perfusion-biotinylated IMCDs yielded 17 positive RT-PCR products from the IMCD suspension (lane +) with 30 cycles of amplification. AQP2 expression served as a positive control for the RT-PCR. PCR without prior RT reaction serves as negative controls and yielded no PCR products (lane −). Together the immunofluorescence staining and RT-PCR confirmed 20 identifications of the 30 selected manually accepted one-peptide identifications.
Fig. 3. Confocal immunofluorescence microscopy confirming the expression of proteins identified from IMCDs.
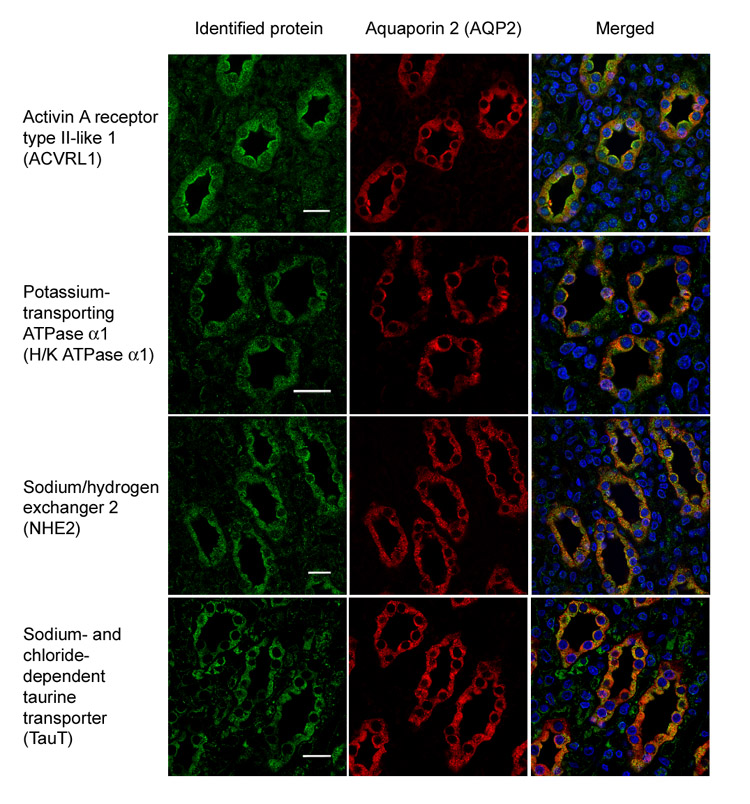
Activin A receptor type II-like I, potassium-transporting ATPase α1, sodium/hydrogen exchanger 2, and sodium- and chloride-dependent taurine transporter in the IMCDs are visualized with secondary antibodies conjugated to FITC or Alexa488 (green). These proteins co-localize with IMCD marker protein aquaporin 2 (red, Alexa568), indicating their expression in the IMCDs. The nuclei are stained with DAPI (blue).
Fig. 4. RT-PCR confirmation of mRNA expression of one-peptide identifications in the IMCDs.
PCR with prior RT reaction (lane +) using AQP2, an IMCD marker protein, specific primers served as a positive control. PCR without reverse transcription reaction (lane −) served as the negative controls. Parentheses indicate sample sources: FPB, fixed perfusion-biotinylated IMCDs; FIB, fixed incubation-biotinylated IMCDs; NPB, non-fixed perfusion-biotinylated IMCDs; NIB, non-fixed incubation-biotinylated IMCDs. ACE, angiotensin-converting enzyme; APC, adenomatous polyposis coli protein; CA4, carbonic anhydrase 4; CD40L, CD40 ligand; CLIC4, chloride intracellular channel protein 4; ERC2, ERC protein 2; FZD1, frizzled 1 precursor; GPR64, G-protein coupled receptor 64 precursor; GRIN2D, glutamate (N-methyl-D-aspartate) receptor subunit ε 4 precursor; HCN1, potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1; Kv4.3, potassium voltage-gated channel subfamily D member 3; LPL, lipoprotein lipase; LRP4, low density lipoprotein receptor-related protein 4; MAPK12, mitogen-activated protein kinase 12; MCT2, monocarboxylate transporter 2; PalmT, palmitoyl-transferase ZDHHC7; PIK3-C2γ, phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing γ polypeptide; PIK3-Cβ, phospha-tidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β isoform; PKA-βC, cAMP-dependent protein kinase, β-catalytic subunit; PODXL, podocalyxin; ROMK, ATP-sensitive inward rectifier potassium channel 1; S1P-R, sphingosine-1-phosphate receptor Edg-8; SCN5A, sodium channel protein type 5 α subunit; SNAP29, synap-tosomal-associated protein 29; THIK-1, potassium channel subfamily K member 13; TYRO3, tyrosine-protein kinase receptor TYRO3; VAPA, vesicle-associated membrane protein-associated protein A; VKγC, vitamin K-dependent gamma-carboxylase; WASPIP, Wiskott-Aldrich syndrome protein interacting protein.
Table II lists 17 integral or GPI-linked membrane proteins that were identified with two or more peptides or with a single peptide and confirmed by immunofluorescence staining or RT-PCR. Type III multiple membrane-spanning helix proteins dominate the list. Note that the hallmark IMCD apical membrane protein AQP2 (1) was not identified in the fixed perfusion-biotinylated IMCDs, consistent with the relative lack of external lysines in this protein (see “Discussion”).
Table II.
Integral and GPI-linked membrane proteins identified from the fixed perfusion-biotinylated inner medullary collecting ducts
| Protein name | Gene name | Accession no. | Ion (n)a | Confirmedb | CDDBc | Typed |
|---|---|---|---|---|---|---|
| Aminopeptidase N | ANPEP | P15684 | 2 (4) | Aminopeptidases | II | |
| Angiotensin-converting enzyme, somatic isoform Precursor | ACE | P47820 | 1 (2) | RT-PCR | Signaling proteins | I |
| ATP-binding cassette subfamily A member 2 | ABCA2 | Q9ESR9 | 2 (3) | Transporters and channels | III | |
| Chloride intracellular channel protein 4 | CLIC4 | Q9Z0W7 | 1 (1) | RT-PCR | Transporters and channels | III |
| G-protein-coupled receptor 64 precursor | GPR64 | Q8CJ11 | 1 (1) | RT-PCR | Membrane receptors | III |
| Lipoprotein lipase precursor | LPL | Q06000 | 1 (1) | RT-PCR | Metabolism proteins | GPI |
| Low density lipoprotein receptor-related protein 4 | LRP4 | Q9QYP1 | 1 (1) | RT-PCR | Membrane receptors | I |
| Palmitoyltransferase ZDHHC7 | ZDHHC7 | Q923G5 | 1 (1) | RT-PCR | Biosynthetic proteins | III |
| Potassium channel subfamily K member 13, THIK-1 | KCNK13 | Q9ERS0 | 1 (1) | RT-PCR | Transporters and channels | III |
| Potassium-transporting ATPase α chain 1 | ATP4A | P09626 | 1 (14) | IF | Ion pumps | III |
| Serine/threonine-protein kinase receptor R3 Precursor | ACVRL1 | P80203 | 2 (5) | Membrane receptors | I | |
| Sodium- and chloride-dependent taurine Transporter | SLC6A6 | P31643 | 1 (3) | IF | Transporters and channels | III |
| Sodium/hydrogen exchanger 2 | SLC9A2 | P48763 | 1 (1) | IF | Transporters and channels | III |
| Syntaxin-8 | STX8 | Q9Z2Q7 | 2 (2) | Membrane trafficking proteins | II | |
| Transmembrane protease, serine 9 | TMPRSS9 | P69526 | 2 (6) | Metabolism proteins | II | |
| Tyrosine-protein kinase receptor TYRO3 precursor | TYRO3 | P55146 | 1 (1) | RT-PCR | Membrane receptors | I |
| Vitamin K-dependent γ -carboxylase | GGCX | O88496 | 1 (1) | RT-PCR | Biosynthetic proteins | III |
Number of peptides identified with their total hit number in parentheses.
Protein identifications confirmed by immunofluorescence (IF) or RT-PCR.
Collecting duct database (CDDB) family group names.
Membrane protein types: I, one membrane-spanning helix with an extracellular N′ terminus; II, one membrane-spanning helix with an extracellular C′ terminus; III, multiple membrane-spanning helices; GPI, glycosylphosphatidylinositol-linked.
Proteomics Analyses of Fixed Incubation-biotinylated IMCDs
We next performed biotinylation in isolated IMCD segments in suspension to selectively label proteins in the basement membrane and the basolateral membrane prior to CaptAvidin affinity protein isolation and LC-MS/MS protein identification. The IMCDs were separated from the non-IMCD structures (loops of Henle and vasculature) of the inner medullae via enzymatic digestion and low speed centrifugation that precipitates the heavier IMCDs (3, 19). After isolation, the IMCD cell integrity was confirmed by the lack of trypan blue permeation (data not shown). The IMCD purity was assessed by immunoblotting (Fig. 5A). As seen, the IMCD suspension (lane I) was enriched in the IMCD marker protein AQP2 and depleted of the non-IMCD marker protein AQP1 compared with the whole inner medulla homogenate (lane W) and the non-IMCD fraction (lane N). When the isolated IMCD suspension was incubated with the biotinylation reagent sulfo-NHS-SS-biotin without prior fixation, biotinylation occurred chiefly at the basement membrane and the basolateral membrane (Fig. 5B, green). For comparison, another IMCD suspension was fixed with 4% paraformaldehyde and biotinylated with sulfo-NHS-LC-biotin. Again the biotinylation occurred at the basement membrane and the basolateral membranes of the IMCD suspension (Fig. 5C, green). The membrane fraction of the fixed incubation-biotinylated IMCD suspension was used for LC-MS/MS protein identification following the same preparation procedures used for the fixed perfusion-biotinylated IMCD sample (Fig. 2A).
Fig. 5. Basolateral surface biotinylation of isolated IMCD suspensions.
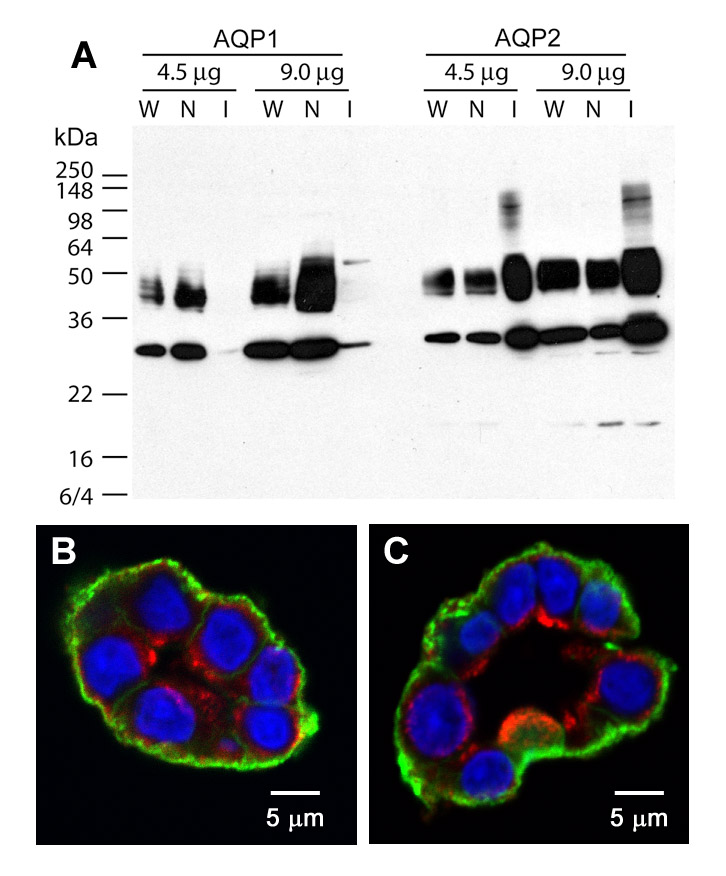
A, an immunoblot shows that the IMCD suspensions (I) are enriched in the IMCD marker protein AQP2 and depleted of the non-IMCD marker protein AQP1 compared with the whole inner medulla homogenate (W) and the non-IMCD suspensions (N). B and C, fluorescence micrographs show that biotinylation occurs at the basement membrane and the basolateral membrane of the isolated IMCD segments in suspensions without (B) or with fixation (C) as revealed by streptavidin-FITC that stains the sites of biotinylation in green. AQP2 antibody identifies IMCD segments (red, Alexa568) and DAPI stains nuclei (blue).
Table III summarizes the numbers of proteins identified in the fixed incubation-biotinylated IMCDs. The computational control experiment indicates the likelihood of a large number of random protein identifications for low numbers of peptide identifications per protein. Consequently we again carried out manual inspection of all spectra to ascertain which identifications were valid. A total of 210 identified proteins are listed in Supplemental Table S-4. Three one-peptide protein identifications were examined with RT-PCR for expression at mRNA level (Fig. 4). 23 integral membrane proteins that were identified with two or more peptides or with one single peptide and confirmed with RT-PCR are presented in Table IV. Type III multiple membrane-spanning helix proteins again dominate the membrane protein identifications. Note that the IMCD basolateral membrane protein AQP4 (23) was not identified in the fixed incubation-biotinylated IMCD suspension due to the lack of external lysine residues (see “Discussion”).
Table III.
Number of proteins identified from the fixed incubation-biotinylated inner medullary collecting ducts
| Peptide numbera | Original database searchb | Reversed database searchc | Net differenced | Manually accepted identificatione |
|---|---|---|---|---|
| 1 | 121 | 25 | 96 | 66 |
| 2 | 53 | 0 | 53 | 44 |
| 3 | 26 | 0 | 26 | 26 |
| 4 | 25 | 0 | 25 | 25 |
| >4 | 50 | 0 | 50 | 49 |
| Total | 275 | 25 | 250 | 210 |
Number of peptides identified for particular proteins.
Number of proteins passing the default criteria of the Bioworks/Sequest search algorithm using the Swiss-Prot database.
Same as Footnote b except the sequences in the database were reversed.
Estimated number of proteins likely to be valid identifications.
Number of proteins passing manual spectrum inspection.
Table IV.
Integral membrane proteins identified from the fixed incubation-biotinylated inner medullary collecting ducts
| Protein name | Gene name | Accession no. | Ion (n)a | Confirmedb | CDDBc | Typed |
|---|---|---|---|---|---|---|
| ADP/ATP translocase 1 | SLC25A4 | Q05962 | 2 (2) | Transporters and channels | III | |
| ADP/ATP translocase 2 | SLC25A5 | Q09073 | 4 (16) | Transporters and channels | III | |
| Agrin precursor | AGRN | P25304 | 4 (20) | Adhesion proteins and collagens | II | |
| Calnexin precursor | CANX | P35565 | 13 (27) | Chaperones | I | |
| CD166 antigen precursor | ALCAM | O35112 | 1 (2) | RT-PCR | Adhesion proteins and collagens | I |
| Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor | COX4I1 | P10888 | 3 (4) | Metabolism proteins | II | |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 63-kDa subunit precursor | RPN2 | P25235 | 5 (8) | Biosynthetic proteins | III | |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 67-kDa subunit precursor | RPN1 | P07153 | 5 (8) | Biosynthetic proteins | III | |
| Epithelial cadherin precursor | CDH1 | Q9R0T4 | 8 (16) | Adhesion proteins and collagens | I | |
| Frizzled 1 precursor | FZD1 | Q08463 | 1 (1) | RT-PCR | Membrane receptors | III |
| Golgi apparatus protein 1 precursor | GLG1 | Q62638 | 2 (4) | Adhesion proteins and collagens | I | |
| Insulin-like growth factor 1 receptor precursor | IGF1R | P24062 | 3 (4) | Membrane receptors | I | |
| Integrin α1 precursor | ITGA1 | P18614 | 2 (2) | Adhesion proteins and collagens | I | |
| Integrin β1 precursor | ITGB1 | P49134 | 24 (54) | Adhesion proteins and collagens | I | |
| Neural cell adhesion molecule L1 precursor | L1CAM | Q05695 | 2 (2) | Adhesion proteins and collagens | I | |
| Neuropilin-1 precursor | NRP1 | Q9QWJ9 | 2 (2) | Membrane receptors | I | |
| Phosphate carrier protein, mitochondrial precursor | SLC25A3 | P16036 | 2 (5) | Transporters and channels | III | |
| Podocalyxin precursor | PODXL | Q9WTQ2 | 1 (1) | RT-PCR | Adhesion proteins and collagens | I |
| Probable G-protein coupled receptor 116 precursor | GPR116 | Q9WVT0 | 2 (4) | Membrane receptors | III | |
| Sodium/potassium-transporting ATPase α-1 chain precursor | ATP1A1 | P06685 | 26 (134) | Ion pumps | III | |
| Sodium/potassium-transporting ATPase α-3 chain | ATP1A3 | P06687 | 15 (63) | Ion pumps | III | |
| Sodium/potassium-transporting ATPase β-1 chain | ATP1B1 | P07340 | 6 (14) | Ion pumps | II | |
| Voltage-dependent anion-selective channel protein 2 | VDAC2 | P81155 | 4 (8) | Transporters and channels | III |
Number of peptides identified with their total hit number in the parentheses.
Protein identifications confirmed by RT-PCR.
Collecting Duct Database (CDDB) family group names.
Membrane protein types: I, one membrane-spanning helix with an extracellular N′ terminus; II, one membrane-spanning helix with an extracellular C′ terminus; III, multiple membrane-spanning helices; GPI, glycosylphosphatidylinositol-linked.
Analyses of Non-integral Membrane Proteins Identified in the Fixed Perfusion-biotinylated and Not in the Incubation-biotinylated IMCDs
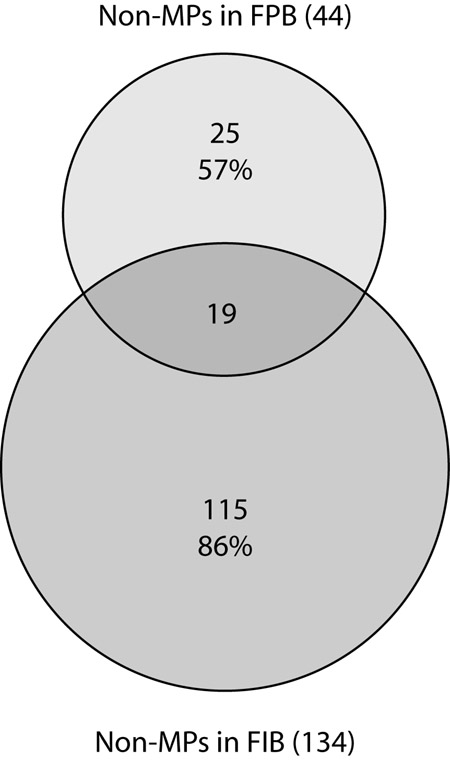
Because our strategy allows identification of integral as well as associated non-integral membrane proteins, the discrete biotinylation of the apical (Fig. 1D) versus the basolateral (Fig. 5C) membrane proteins allows identification of the non-integral membrane proteins from respective membrane protein domains. Apically associated non-integral membrane proteins are those identified in the fixed perfusion-biotinylated IMCDs. Basolaterally associated non-integral membrane proteins are those identified in the fixed incubation-biotinylated IMCD suspension. Fig. 6 shows a general comparison of proteins from the two samples identified with two or more peptides or with a single peptide and confirmed with RT-PCR. As seen (Fig. 6), the majority of the non-integral membrane proteins identified in the fixed perfusion-biotinylated IMCDs (25 of 44 or 57%) do not overlap with those identified in the fixed incubation-biotinylated IMCD suspension. These 25 non-integral membrane proteins identified exclusively in the fixed perfusion-biotinylated IMCDs are listed in Table V and include a number of proteins involved in cell signaling, vesicular trafficking, and cytoskeletal organization. They can be considered potential components of the vasopressin-regulated apical trafficking mechanisms. However, this subtractive comparison of the non-integral membrane proteins between the fixed perfusion- and incubation-biotinylated IMCDs cannot be used to exclude the existence of a particular protein in either apical or basolateral membrane domains due to the limited sensitivity of the methods.
Fig. 6. A Venn diagram summarizing the non-integral membrane proteins (non-MPs) identified in the fixed perfusion-biotinylated (FPB) IMCD and those identified in the fixed incubation-biotinylated (FIB) IMCD suspension.
Numbers indicate protein identifications. Percentages indicate non-overlapping identifications in a particular sample preparation.
Proteomics Analyses of Non-fixed Perfusion- and Incubation-biotinylated IMCDs
Without fixation, biotinylation occurs throughout the IMCD cells but nevertheless is limited to IMCD cells (Fig. 1C) and therefore provides a means for general identification of IMCD proteins. Similarly the high purity of the isolated IMCD suspension also permits valid IMCD protein identifications (Fig. 5A). Therefore, samples from the non-fixed perfusion- and incubation-biotinylated (using sulfo-NHS-SS-biotin) were isolated with streptavidin-agarose affinity chromatography and analyzed with mass spectrometry. The computational control experiments and the results of manual inspection are summarized in Supplemental Table S-5. Specific proteins from non-fixed perfusion- and incubation-biotinylated IMCDs are combined in Supplemental Table S-6. 34 integral and GPI-linked membrane proteins that were identified with two or more peptides or with a single peptide and confirmed with RT-PCR (Fig. 4) are presented in Table VI. Type III membrane proteins are again the dominant membrane proteins. Note that six GPI-linked proteins were identified in the non-fixed perfusion-biotinylated IMCDs. Also the perfusion-biotinylation without prior fixation led to the identifications of apical (AQP2) and basolateral membrane proteins (AQP4, barttin, and E-cadherin), indicating the entry of the biotinylation reagents. Likewise the incubation-biotinylation without prior fixation led to the identification of the apical (AQP2) and basolateral (AQP4) membrane proteins.
Table VI.
Integral and GPI-linked membrane proteins identified in the non-fixed perfusion- and incubation-biotinylated inner medullary collecting ducts
| Protein name | Gene name | Accession no. | Ion (n)a | Confirmedb | CDDBc | Typed | P/Ic |
|---|---|---|---|---|---|---|---|
| ADP/ATP translocase 1 | SLC25A4 | Q05962 | 2 (2) | Transporters and channels | III | I | |
| ADP/ATP translocase 2 | SLC25A5 | Q09073 | 4 (16) | Transporters and channels | III | I | |
| Aquaporin-1 | AQP1 | P29975 | 4 (5) | Transporters and channels | III | I | |
| Aquaporin-2 | AQP2 | P34080 | 3 (7) | Transporters and channels | III | P, I | |
| Aquaporin-4 | AQP4 | P47863 | 3 (4) | Transporters and channels | III | P, I | |
| Barttin | BSND | Q8R2H3 | 7 (47) | Transporters and channels | III | P | |
| Basigin precursor | BSG | P26453 | 2 (2) | Transporters and channels | I | I | |
| Calnexin precursor | CANX | P35565 | 13 (27) | Chaperones | I | P, I | |
| Carbonic anhydrase 4 precursor | CA4 | P48284 | 1 (4) | RT-PCR | Transporters and channels (related) | GPI | P |
| Carnitine O-palmitoyltransferase I, liver isoform | CPT1A | P32198 | 3 (4) | Metabolism proteins | III | I | |
| CD166 antigen precursor | ALCAM | O35112 | 1 (2) | RT-PCR | Adhesion proteins and collagens | I | I |
| CD48 antigen precursor | CD48 | P10252 | 3 (4) | Signaling proteins | GPI | P | |
| CD59 glycoprotein precursor | CD59 | P27274 | 1 (5) | RT-PCR | Miscellaneous proteins | GPI | P |
| Chloride channel protein ClC-Ka | CLCNKA | Q06393 | 2 (3) | Transporters and channels | III | I | |
| Contactin-1 precursor | CNTN1 | Q63198 | 2 (2) | Signaling proteins | GPI | P | |
| Dihydropyridine-sensitive L- type, calcium channel α-2/δ subunits precursor | CACNA2D1 | P54290 | 7 (24) | Transporters and channels | III | P | |
| Epithelial cadherin precursor | CDH1 | Q9R0T4 | 5 (78) | Adhesion proteins and collagens | I | P | |
| Integrin α1 precursor | ITGA1 | P18614 | 2 (2) | Adhesion proteins and collagens | I | I | |
| Mitochondrial 2-oxoglutarate/malate carrier protein | SLC25A11 | P97700 | 2 (3) | Transporters and channels | III | I | |
| Monocarboxylate transporter 2, MCT2 | SLC16A7 | Q63344 | 1 (2) | RT-PCR | Transporters and channels | III | I |
| Monocyte differentiation antigen CD14 precursor | CD14 | Q63691 | 3 (6) | Signaling proteins | GPI | P | |
| Phosphate carrier protein, mitochondrial precursor | SLC25A3 | P16036 | 2 (5) | Transporters and channels | III | I | |
| Podocalyxin precursor | PODXL | Q9WTQ2 | 1 (1) | RT-PCR | Adhesion proteins and collagens | I | I |
| Potassium-transporting ATPase α chain 2 | ATP12A | P54708 | 2 (3) | Ion pumps | III | I | |
| Reticulon-4 | RTN4 | Q9JK11 | 8 (24) | Membrane trafficking proteins | III | I | |
| Sideroflexin-3 | SFXN3 | Q9JHY2 | 3 (4) | Transporters and channels | III | I | |
| Sodium/potassium-transporting ATPase α-1 chain precursor | ATP1A1 | P06685 | 26 (134) | Ion pumps | III | I | |
| Sodium/potassium-transporting ATPase α-3 chain | ATP1A3 | P06687 | 15 (63) | Ion pumps | III | I | |
| Sodium/potassium-transporting ATPase α-4 chain | ATP1A4 | Q64541 | 2 (3) | Ion pumps | III | I | |
| Sodium/potassium-transporting ATPase β-1 chain | ATP1B1 | P07340 | 6 (14) | Ion pumps | II | I | |
| Thy-1 membrane glycoprotein precursor | THY1 | P01830 | 2 (5) | Adhesion proteins and collagens | GPI | P | |
| Vesicle-associated membrane protein-associated protein A | VAPA | Q9Z270 | 1 (1) | RT-PCR | Membrane trafficking proteins | II | I |
| Voltage-dependent anion-selective channel protein 1 | VDAC1 | Q9Z2L0 | 7 (23) | Transporters and channels | III | P, I | |
| Voltage-dependent anion-selective channel protein 2 | VDAC2 | P81155 | 4 (8) | Transporters and channels | III | I |
Number of peptides identified with their total hit number in the parentheses.
Protein identifications confirmed by RT-PCR.
Collecting Duct Database (CDDB) family group names.
Membrane protein types: I, one membrane-spanning helix with an extracellular N′ terminus; II, one membrane-spanning helix with an extracellular C′ terminus; III, multiple membrane-spanning helices; GPI, glycosylphosphatidylinositol-linked.
Protein identified with perfusion (P) or incubation (I) biotinylation method.
IMCD Membrane Protein Database
Overall we identified 62 integral and GPI-linked IMCD membrane proteins in this study. These proteins together with previously identified membrane proteins (n = 187) were used to populate a new IMCD Membrane Protein (IMP) Database (dir.nhlbi.nih.gov/papers/lkem/imp/index.htm). Note that 27 of the membrane proteins identified in this study were also among the 187 found in our prior studies, giving a total of 222 proteins in the database.
DISCUSSION
The chief objective of this study was to expand the identified proteome of renal inner medullary collecting duct cells through use of surface biotinylation to enrich plasma membrane proteins. Overall we expanded the current IMCD Pro-teome Database by 35 integral and GPI-linked membrane proteins, and we introduce a new database of integral membrane proteins and GPI-linked proteins (dir.nhlbi.nih.gov/papers/lkem/imp/index.htm). In addition, we exploited the surface biotinylation technique to differentially label and enrich apical and basolateral membrane proteins, thereby allowing analyses of respective membrane proteomes. Beyond these membrane proteins, subtractive comparison of the non-integral membrane proteins identified in the fixed perfusion- and not in the incubation-biotinylated IMCDs revealed a number of proteins that are potentially involved in vasopressin signaling and AQP2 trafficking. Many of these non-integral membrane proteins were likely to be present as a result of physical association (i.e. binding) with the labeled integral membrane proteins, although with the current data we cannot infer the presence of such an association for any particular protein.
Surface biotinylation is a rather popular technique for studying plasma membrane proteins (24). However, several technical pitfalls have been reported regarding the labeling chemistry and efficiency of the biotinylation reagents (25) as well as the isolation of the biotinylated proteins using strepta-vidin affinity chromatography (20). In this study, we discovered an additional limitation of this technique, namely that biotinylation reagents, including so-called membrane-impermeant forms, can enter cells even at 2 °C at which endocy-tosis can be expected to be effectively inhibited. Thus, when inner medullary collecting ducts were perfused with sulfo-NHS-SS-biotin to label proteins of the apical plasma membrane (Fig. 1, A and B), we found by streptavidin-FITC labeling of tissue sections that cytoplasmic proteins were labeled in the perfused IMCD cells even at 2 °C (Fig. 1C). Another similar reagent, sulfo-NHS-LC-biotin, led to the same result, indicating a common problem with this type of biotinylation reagents. A determination of the mechanism of entry of the biotinylation reagent is beyond the scope of this study. It is possible that lowering the temperature to 2 °C does not completely inhibit endocytosis or metabolic activities. Remarkably renal tubules were reported to maintain active transport functions at 0 °C (26), and other ATP-dependent activities seem possible. Alternatively the reagents may enter the cells through endocytosis-independent mechanisms similar to those responsible for cellular entry of certain cell-penetrating peptides (27).
The intracellular biotinylation was prevented when preceded by a brief fixation with paraformaldehyde (Fig. 1, C and D). Presumably the intracellular biotinylation was prevented because paraformaldehyde cross-links unsaturated fatty acids and proteins in the plasma membrane (16), forming a rigid shield on the cell surface resistant to internalization into the IMCD cells at 2 °C. Although the internalization of the biotinylation reagents from the basolateral membrane into the cells did not appear to be as extensive as that from the apical membrane (Figs. 5B and 1C), the identification of both apical (AQP2) and basolateral (AQP4) membrane proteins in the non-fixed incubation-biotinylated IMCDs (Table VI) indicates the entry of the biotinylation reagents (see below). Overall perfusion and incubation-biotinylation with prior fixation allowed labeling, enrichment, and analyses, respectively, of apical and basolateral IMCD membrane proteomes (Tables II and IV). Perfusion- and incubation-biotinylation without prior fixation permitted valid identification of IMCD proteins (Table VI) because of the confined labeling in the perfused IMCD cells (Fig. 1C) and the high purity of the IMCD suspension (Fig. 5A).
The membrane proteins identified in the non-fixed and fixed biotinylated IMCDs have little overlap (Tables II, IV, and VI). We categorized the membrane proteins according to their types (28) to learn why some proteins were preferentially identified in the fixed versus the non-fixed perfusion-biotinylated IMCDs (Fig. 7). Type I and II membrane proteins have one single transmembrane span with N′ or C′ terminus facing the extracellular space, respectively. Type III membrane proteins contain multiple membrane-spanning α helices. Other proteins are anchored to the membrane via a GPI anchor. Fig. 7 shows some examples of membrane protein types identified in the fixed (top panel) and the non-fixed (bottom panel) perfusion-biotinylated IMCDs. In general, all four types of membrane proteins were identified in either sample preparation. The peptides identified (Fig. 7, red) by the methods used in this study were limited to the N′ and C′ termini and the loops that connect membrane-spanning domains of the membrane proteins. In the fixed perfusion-biotinylated IMCDs where the biotinylation reagents were restricted to the luminal surface of IMCDs, type III polytopic membrane proteins (e.g. NHE2) were the predominant membrane proteins identified (Table II) most likely because these membrane proteins contain more extracellular lysines accessible to the biotinylation reagents. Similarly the type I and II membrane proteins (low density lipoprotein receptor-related protein 4, ACVRL1, and aminopeptidase N) identified in the fixed perfusion-biotinylated IMCDs contain several extracellularly accessible lysines (Fig. 7). AQP2 has only one extracellular lysine, which presumably is not accessible to the biotinylation reagent and therefore explains why this hallmark IMCD apical membrane protein was not identified in the fixed perfusion-biotinylated IMCDs. AQP2, however, was identified in the non-fixed perfusion-biotinylated IMCDs (Table VI) where the biotinylation reagents were able to penetrate the apical membrane (Fig. 1C) and react with three lysines on the cytosolic C′ terminus of AQP2 in the apical membrane or in intracellular vesicles. Some basolateral membrane proteins including E-cadherin (29), barttin (30), and AQP4 (23) were identified in the non-fixed perfusion-biotinylated IMCDs (Table VI) likely through the same mechanism. Similarly the basolateral membrane protein AQP4, which lacks extracellular lysine, was not identified in the fixed incubation-biotinylated IMCD but was identified in the non-fixed incubation-biotinylated IMCD suspension (Table VI) as the reagents enter the cells. The entering biotinylation reagent from the non-fixed basolateral membrane also led to the identification of the apical membrane protein AQP2 (Table VI).
Fig. 7. Examples of integral and GPI-linked membrane proteins identified in the fixed (top) and the non-fixed (bottom) perfusion-biotinylated IMCDs.
Type I and II membrane proteins have one single transmembrane span with N′ or C′ terminus facing the extracellular space, respectively. Type III membrane proteins contain multiple membrane-spanning topology. Some membrane proteins are anchored to the membrane via a GPI anchor. LPL, lipoprotein lipase; LRP4, low density lipoprotein receptor-related protein 4; ANPEP, aminopeptidase N.
Although paraformaldehyde effectively inhibited the entry of the biotinylation reagents, its use comes with two major drawbacks. First, paraformaldehyde reacts with primary amines at the N′ termini and lysine residues needed for biotinylation, leading to low protein yield and decreased signal to noise ratio for protein identification by LC-MS/MS. Second, both paraformaldehyde and the biotinylation reagents modify lysines that define sites of peptide bond hydrolysis by trypsin. These factors combine to decrease the sensitivity of the overall approach. In particular, this low sensitivity combines with the low sensitivity intrinsic to the IMCD perfusion approach as a means of labeling apical proteins, diminishing prospects for high numbers of multiple-peptide identifications of membrane proteins. These circumstances point to the need to carry through with immunological or RT-PCR identification of the most interesting single-peptide identifications.
Despite the low sensitivity, fixation prior to biotinylation permitted differential labeling and enrichment of apical and basolateral IMCD membrane proteins, which are reflected in the respective proteomes identified by the LC-MS/MS. For example, the fixed perfusion-biotinylation method labeled only the apical membrane of IMCDs (Fig. 1D) and led to the identification of two known apical membrane proteins, H+/K+-ATPase α1 (31) and NHE2 (32). The fixed incubation-biotinylation method labeled only the basolateral membrane of IMCDs (Fig. 5C) and led to the identifications of known basolateral membrane proteins including Na+/K+-ATPase (33) and E-cadherin (29).
Among the membrane proteins identified from the fixed perfusion-biotinylated IMCDs, ACRVL1, H+/K+-ATPase α1, NHE2, and TauT were confirmed to be present in the IMCDs with immunofluorescence staining and confocal microscopy (Fig. 3). The staining of ACVRL1 showed apical localization. ACVRL1, also known as transforming growth factor-β receptor type I, is of interest because transforming growth factor-β antagonizes the effects of aldosterone in IMCD cells (34). In addition, activin A together with Wnt4 and hepatocyte growth factor is involved in promoting renal tubule formation and AQP2 expression (35). The staining of H+/K+-ATPase α1 and NHE2 was consistent with their apical localization (31, 32). The presence of an H+/K+-ATPase in collecting duct cells has been recognized based on immunohistochemical and functional studies, although the isoform (colonic versus gastric) has been controversial. Based on the high quality of the spectra, we conclude here that the gastric isoform is expressed in the rat IMCD, although this identification does not exclude the presence of the colonic isoform as well. TauT, a sodium- and chloride-dependent taurine transporter, is an interesting protein because mice lacking TauT have impaired ability to increase water excretion in response to water loading (36). Another potential apical membrane marker protein is the GPI-linked membrane protein lipoprotein lipase (Table II). GPI-linked proteins are targeted to the apical membrane of most cells (37). It is interesting to note that six other GPI-linked proteins were identified in the non-fixed perfusion-bio-tinylated IMCDs. Of them, CD59 (38) and carbonic anhydrase IV (39) have been shown to be expressed in apical membrane of human and rabbit IMCDs.
The discrete biotinylation of the apical and the basolateral membranes allowed subtractive analysis of non-integral membrane proteins identified in the fixed perfusion-biotinylated IMCDs and not in the fixed incubation-biotinylated IM-CDs. This subtractive comparison cannot be used to exclude the existence of a particular protein in either apical or basolateral membrane compartment due to the limited sensitivity of the overall approach. It only generates potential hypotheses for further experimentation. At least two of the proteins that were identified, cAMP-dependent kinase catalytic subunit and Ca2+/calmodulin-dependent nitric-oxide synthase 1, are likely involved in the signaling network associated with vasopressin action in the IMCD (40). Another identified protein, calcyclin, a calcium-binding protein, is proposed to act as a transducing molecule that couples a vasopressin stimulus to AQP2 trafficking through its interaction with annexins and actin-binding proteins such as caldesmon, tropomyosin, and calponin (41). Some scaffold proteins including bassoon, piccolo, and septin 9 involved in cytoskeleton and membrane organization as well as synaptic vesicle trafficking are of potential interest because vasopressin-stimulated AQP2 trafficking involves cytoskeletal reorganization (42). In particular, septin 9 was recently identified in IMCDs as a protein phosphorylated at threonine residues (13), and septin polymerization is regulated by phosphorylation and small GTP-binding proteins (43). Moreover septin 9 associates with SNARE proteins suggesting its role in vesicle docking (43). The identification of a small GTP-binding protein (Rab31), Rab-interacting proteins (granuphilin and Rabaptin-5), and a Rap1 GTPase-activating protein (signal-induced proliferation-associated 1-like protein 1 (SPA-1)) together with cytoskeleton proteins and molecular motor is potentially relevant to the mechanism of AQP2 trafficking. In fact, SPA-1 has previously been implicated in AQP2 trafficking (44).
Supplementary Material
Table V.
Non-integral membrane proteins identified in the fixed perfusion-biotinylated and not in the fixed incubation-biotinylated inner medullary collecting ducts
| Protein namea | Gene name | Accession no. | Ion (n)b | Confirmedc | Functional remark |
|---|---|---|---|---|---|
| Alcohol dehydrogenase | AKR1A1 | P51635 | 2 (2) | Metabolism proteins. | |
| α 1-Antiproteinase precursor | SERPINA1 | P17475 | 10 (12) | Secreted protein. | |
| Bassoon protein | BSN | O88778 | 2 (4) | Scaffold protein; myristoylated; forms complex with ERC2; similar to piccolo; membrane-associated; organization of the cytomatrix at the nerve terminal active zone. | |
| Calcyclin | S100A6 | P05964 | 1 (1) | RT-PCR | Signaling protein; regulated by vasopressin (41); binds to prolactin receptor; binds to annexins. |
| cAMP-dependent protein kinase, β-catalytic subunit | PRKACB | P68182 | 1 (3) | RT-PCR | Protein kinase, serine/threonine; may be membrane-associated via A kinase anchoring proteins. |
| Creatine kinase, sarcomeric mitochondrial precursor | CKMT2 | P09605 | 3 (12) | Metabolism protein; thought to be found only in heart and skeletal muscles. | |
| Glandular kallikrein-7, submandibular/renal precursor | KLK7 | P36373 | 2 (4) | Protein degradation pathway; predominant kallikrein protein in the kidney; membrane-associated and secreted protein. | |
| Glial fibrillary acidic protein, astrocyte | GFAP | P47819 | 3 (77) | Cytoskeletal proteins. | |
| Hermansky-Pudlak syndrome 6 protein homolog | HPS6 | Q7M733 | 2 (2) | Miscellaneous proteins. | |
| Myosin-5A | MYO5A | Q9QYF3 | 3 (10) | Motor protein; processive actin-based motor; membrane-associated; involved in vesicle transport along actin filaments. | |
| Nitric-oxide synthase 1, brain | NOS1 | P29476 | 3 (10) | Signaling protein; produces NO from arginine; stimulated by calcium/calmodulin. | |
| PDZ domain-containing RING finger protein 3 | PDZRN3 | P68907 | 3 (4) | Miscellaneous proteins. | |
| Peroxiredoxin-2 | PRDX2 | P35704 | 2 (2) | Stress protein; redox regulation in cell; role in eliminating peroxides generated during metabolism. | |
| Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β isoform | PIK3CB | Q9Z1L0 | 1 (1) | RT-PCR | Phosphoinositide 3-kinase; peripheral membrane protein. |
| Piccolo protein | PCLO | Q9JKS6 | 2 (8) | Scaffold protein; similar to bassoon; membrane-associated; organization of the cytomatrix at the nerve terminal active zone; binds profilin. | |
| Propionyl-CoA carboxylase α chain, mitochondrial precursor | PCCA | P14882 | 6 (11) | Metabolism protein; key enzyme in the catabolic pathway of odd-chain fatty acids, isoleucine, threonine, methionine, and valine. | |
| Rab GTPase-binding effector protein 1 | RABEP1 | O35550 | 1 (2) | RT-PCR | Small GTP-binding protein-related protein; Rab effector protein acting as linker between γ-adaptin, Rab4A, and Rab5A; involved in endocytic membrane fusion and membrane trafficking of recycling endosomes; stimulates RabGEF1-mediated nucleotide exchange on Rab5A. |
| Ras-related protein Rab-31 | RAB31 | Q6GQP4 | 1 (4) | RT-PCR | Small GTP-binding proteins. |
| Septin-9 | SEPT9 | Q9QZR6 | 1 (4) | RT-PCR | Scaffold protein; binds GTP; associates with cell membranes and the actin and microtubule cytoskeletons; polymerize to form filamentous structures under the influence of Rho and CDC42; regulated by phosphorylation; associate with SNARE complexes (43). |
| SPA-1 | SIPA1L1 | O35412 | 2 (2) | Small GTP-binding protein-related protein; Rap1GAP; implicated in regulation of AQP2 trafficking (44). | |
| Synaptosomal-associated protein 29 | SNAP29 | Q9Z2P6 | 1 (1) | RT-PCR | SNARE protein. |
| Synaptotagmin-like protein 4 (granuphilin) | SYTL4 | Q8VHQ7 | 3 (9) | Small GTP-binding protein-related protein; peripheral membrane protein; part of a ternary complex containing STX1A and RAB27A; interacts with syntaxins in munc18-depenedent manner; may play a role in vesicle trafficking. | |
| Timeless homolog | TIMELESS | Q9Z2Y1 | 2 (2) | Nuclear protein; play a role in epithelial morphogenesis during kidney development and branching of tubules (45). | |
| Wiskott-Aldrich syndrome protein-interacting protein | WASPIP | Q6IN36 | 1 (1) | RT-PCR | Scaffold protein; induces actin polymerization and redistribution; binds WASP, profilin, actin, and GRB2. |
Non-integral membrane proteins identified in the fixed incubation-biotinylated IMCDs subtracted from those identified in the fixed perfusion-biotinylated IMCDs.
Number of peptides identified with their total hit number in the parentheses.
Identifications confirmed by RT-PCR.
Acknowledgments
We thank Zu-Xi Yu (Pathology Core Facility, NHLBI, National Institutes of Health) as well as Christian A. Combs and Daniela Malide (Light Microscopy Core Facility, NHLBI, National Institutes of Health) for professional skills and advice regarding confocal fluorescence microscopy.
Footnotes
This work was supported by the intramural budget of NHLBI, National Institutes of Health Grant Z01-HL-01285.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental materials.
The abbreviations used are: ENaC, epithelial sodium channel; ACVRL1, activin A receptor type-II like I; AQP, aquaporin; DAPI, 4′,6-diamidino-2-phenylindole; GPI, glycosylphosphatidylinositol; H+/K+-ATPase, potassium-transporting ATPase; IMCD, inner medullary collecting duct; Na+/K+-ATPase, sodium/potassium-transporting ATPase; NHE2, sodium/hydrogen exchanger 2; SPA-1, signal-induced proliferation-associated 1-like protein 1; TauT, sodium- and chloride-dependent taurine transporter; UT-A, urea transporter A; Wnt4, wingless-type mouse mammary tumor virus integration site family member 4; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; NHS, N-hydroxysuccinimide; FD&C, United States Federal Food, Drug, and Cosmetic Act.
References
- 1.Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci USA. 1995;92:1013–1017. doi: 10.1073/pnas.92.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiGiovanni SR, Nielsen S, Christensen EI, Knepper MA. Regulation of collecting duct water channel expression by vasopressin in Brattleboro rat. Proc Natl Acad Sci USA. 1994;91:8984–8988. doi: 10.1073/pnas.91.19.8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ecelbarger CA, Terris J, Frindt G, Echevarria M, Marples D, Nielsen S, Knepper MA. Aquaporin-3 water channel localization and regulation in rat kidney. Am J Physiol. 1995;269:F663–F672. doi: 10.1152/ajprenal.1995.269.5.F663. [DOI] [PubMed] [Google Scholar]
- 4.Ecelbarger CA, Kim GH, Terris J, Masilamani S, Mitchell C, Reyes I, Verbalis JG, Knepper MA. Vasopressin-mediated regulation of epithelial sodium channel abundance in rat kidney. Am J Physiol. 2000;279:F46–F53. doi: 10.1152/ajprenal.2000.279.1.F46. [DOI] [PubMed] [Google Scholar]
- 5.Nicco C, Wittner M, DiStefano A, Jounier S, Bankir L, Bouby N. Chronic exposure to vasopressin upregulates ENaC and sodium transport in the rat renal collecting duct and lung. Hypertension. 2001;38:1143–1149. doi: 10.1161/hy1001.092641. [DOI] [PubMed] [Google Scholar]
- 6.Sands JM, Nonoguchi H, Knepper MA. Vasopressin effects on urea and H2O transport in inner medullary collecting duct subsegments. Am J Physiol. 1987;253:F823–F832. doi: 10.1152/ajprenal.1987.253.5.F823. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Sands JM, Klein JD. Vasopressin rapidly increases phosphorylation of UT-A1 urea transporter in rat IMCDs through PKA. Am J Physiol. 2002;282:F85–F90. doi: 10.1152/ajprenal.0054.2001. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa SE, Schrier RW. Pathophysiological roles of arginine vasopressin and aquaporin-2 in impaired water excretion. Clin Endocrinol (Oxf) 2003;58:1–17. doi: 10.1046/j.1365-2265.2003.01647.x. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen S, Frokiaer J, Marples D, Kwon TH, Agre P, Knepper MA. Aquaporins in the kidney: from molecules to medicine. Physiol Rev. 2002;82:205–244. doi: 10.1152/physrev.00024.2001. [DOI] [PubMed] [Google Scholar]
- 10.Hoffert JD, van Balkom BW, Chou CL, Knepper MA. Application of difference gel electrophoresis to the identification of inner medullary collecting duct proteins. Am J Physiol. 2004;286:F170–F179. doi: 10.1152/ajprenal.00223.2003. [DOI] [PubMed] [Google Scholar]
- 11.van Balkom BW, Hoffert JD, Chou CL, Knepper MA. Proteomic analysis of long-term vasopressin action in the inner medullary collecting duct of the Brattleboro rat. Am J Physiol. 2004;286:F216–F224. doi: 10.1152/ajprenal.00307.2003. [DOI] [PubMed] [Google Scholar]
- 12.Hoorn EJ, Pisitkun T, Zietse R, Gross P, Frokiaer J, Wang NS, Gonzales PA, Star RA, Knepper MA. Prospects for urinary proteomics: exosomes as a source of urinary biomarkers. Nephrology (Carlton) 2005;10:283–290. doi: 10.1111/j.1440-1797.2005.00387.x. [DOI] [PubMed] [Google Scholar]
- 13.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA. 2006;103:7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutillas PR, Biber J, Marks J, Jacob R, Stieger B, Cramer R, Waterfield M, Burlingame AL, Unwin RJ. Proteomic analysis of plasma membrane vesicles isolated from the rat renal cortex. Proteomics. 2005;5:101–112. doi: 10.1002/pmic.200400886. [DOI] [PubMed] [Google Scholar]
- 15.Sands JM, Knepper MA. Urea permeability of mammalian inner medullary collecting duct system and papillary surface epithelium. J Clin Investig. 1987;79:138–147. doi: 10.1172/JCI112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D. Reactions of aldehydes with unsaturated fatty acids during histological fixation. Histochem J. 1972;4:421–465. doi: 10.1007/BF01012533. [DOI] [PubMed] [Google Scholar]
- 17.Chou CL, DiGiovanni SR, Luther A, Lolait SJ, Knepper MA. Oxytocin as an antidiuretic hormone. II. Role of V2 vasopressin receptor. Am J Physiol. 1995;269:F78–F85. doi: 10.1152/ajprenal.1995.269.1.F78. [DOI] [PubMed] [Google Scholar]
- 18.Inoue T, Terris J, Ecelbarger CA, Chou CL, Nielsen S, Knepper MA. Vasopressin regulates apical targeting of aquaporin-2 but not of UT1 urea transporter in renal collecting duct. Am J Physiol. 1999;276:F559–F566. doi: 10.1152/ajprenal.1999.276.4.F559. [DOI] [PubMed] [Google Scholar]
- 19.Marples D, Knepper MA, Christensen EI, Nielsen S. Redistribution of aquaporin-2 water channels induced by vasopressin in rat kidney inner medullary collecting duct. Am J Physiol. 1995;269:C655–C664. doi: 10.1152/ajpcell.1995.269.3.C655. [DOI] [PubMed] [Google Scholar]
- 20.Morag E, Bayer EA, Wilchek M. Reversibility of biotin-binding by selective modification of tyrosine in avidin. Biochem J. 1996;316:193–199. doi: 10.1042/bj3160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barile M, Pisitkun T, Yu MJ, Chou CL, Verbalis MJ, Shen RF, Knepper MA. Large scale protein identification in intracellular aquaporin-2 vesicles from renal inner medullary collecting duct. Mol Cell Proteomics. 2005;4:1095–1106. doi: 10.1074/mcp.M500049-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terris J, Ecelbarger CA, Marples D, Knepper MA, Nielsen S. Distribution of aquaporin-4 water channel expression within rat kidney. Am J Physiol. 1995;269:F775–F785. doi: 10.1152/ajprenal.1995.269.6.F775. [DOI] [PubMed] [Google Scholar]
- 24.Gauthier DJ, Gibbs BF, Rabah N, Lazure C. Utilization of a new biotinylation reagent in the development of a nondiscriminatory investigative approach for the study of cell surface proteins. Proteomics. 2004;4:3783–3790. doi: 10.1002/pmic.200300860. [DOI] [PubMed] [Google Scholar]
- 25.Gottardi CJ, Dunbar LA, Caplan MJ. Biotinylation and assessment of membrane polarity: caveats and methodological concerns. Am J Physiol. 1995;268:F285–F295. doi: 10.1152/ajprenal.1995.268.2.F285. [DOI] [PubMed] [Google Scholar]
- 26.Burg MB, Orloff J. Active cation transport by kidney tubules at 0 C. Am J Physiol. 1964;207:983–988. doi: 10.1152/ajplegacy.1964.207.5.983. [DOI] [PubMed] [Google Scholar]
- 27.Noguchi H, Matsumoto S. Protein transduction technology: a novel therapeutic perspective. Acta Med Okayama. 2006;60:1–11. doi: 10.18926/AMO/30757. [DOI] [PubMed] [Google Scholar]
- 28.Cai YD, Chou KC. Predicting membrane protein type by functional domain composition and pseudo-amino acid composition. J Theor Biol. 2006;238:395–400. doi: 10.1016/j.jtbi.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 29.McDill BW, Li SZ, Kovach PA, Ding L, Chen F. Congenital progressive hydronephrosis (cph) is caused by an S256L mutation in aquaporin-2 that affects its phosphorylation and apical membrane accumulation. Proc Natl Acad Sci USA. 2006;103:6952–6957. doi: 10.1073/pnas.0602087103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estevez R, Boettger T, Stein V, Birkenhager R, Otto E, Hildebrandt F, Jentsch TJ. Barttin is a Cl− channel β-subunit crucial for renal Cl− reabsorption and inner ear K+ secretion. Nature. 2001;414:558–561. doi: 10.1038/35107099. [DOI] [PubMed] [Google Scholar]
- 31.Wingo CS, Smolka AJ. Function and structure of H-K-ATPase in the kidney. Am J Physiol. 1995;269:F1–F16. doi: 10.1152/ajprenal.1995.269.1.F1. [DOI] [PubMed] [Google Scholar]
- 32.Chambrey R, Warnock DG, Podevin RA, Bruneval P, Mandet C, Belair MF, Bariety J, Paillard M. Immunolocalization of the Na+/H+ exchanger isoform NHE2 in rat kidney. Am J Physiol. 1998;275:F379–F386. doi: 10.1152/ajprenal.1998.275.3.F379. [DOI] [PubMed] [Google Scholar]
- 33.Kashgarian M, Biemesderfer D, Caplan M, Forbush B., III Monoclonal antibody to Na, K-ATPase: immunocytochemical localization along nephron segments. Kidney Int. 1985;28:899–913. doi: 10.1038/ki.1985.216. [DOI] [PubMed] [Google Scholar]
- 34.Stokes JB. Physiologic resistance to the action of aldosterone. Kidney Int. 2000;57:1319–1323. doi: 10.1046/j.1523-1755.2000.00969.x. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi T, Tanaka H, Kuwana H, Inoshita S, Teraoka H, Sasaki S, Terada Y. Wnt4-transformed mouse embryonic stem cells differentiate into renal tubular cells. Biochem Biophys ResCommun. 2005;336:585–595. doi: 10.1016/j.bbrc.2005.08.136. [DOI] [PubMed] [Google Scholar]
- 36.Huang DY, Boini KM, Lang PA, Grahammer F, Duszenko M, Heller-Stilb B, Warskulat U, Haussinger D, Lang F, Vallon V. Impaired ability to increase water excretion in mice lacking the taurine transporter gene TAUT. Pfluegers Arch Eur J Physiol. 2006;451:668–677. doi: 10.1007/s00424-005-1499-y. [DOI] [PubMed] [Google Scholar]
- 37.Polishchuk R, Di Pentima A, Lippincott-Schwartz J. Delivery of raft-associated, GPI-anchored proteins to the apical surface of polarized MDCK cells by a transcytotic pathway. Nat Cell Biol. 2004;6:297–307. doi: 10.1038/ncb1109. [DOI] [PubMed] [Google Scholar]
- 38.Bridgewater DJ, Mok A, Matsell DG. Expression of complement regulatory proteins in the developing human kidney. Pediatr Nephrol. 2000;15:36–42. doi: 10.1007/s004670000390. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz GJ, Kittelberger AM, Barnhart DA, Vijayakumar S. Carbonic anhydrase IV is expressed in H(+)-secreting cells of rabbit kidney. Am J Physiol. 2000;278:F894–F904. doi: 10.1152/ajprenal.2000.278.6.F894. [DOI] [PubMed] [Google Scholar]
- 40.Pisitkun T, Bieniek J, Tchapyjnikov D, Wang G, Wu WW, Shen RF, Knepper MA. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol Genomics. 2006;25:263–276. doi: 10.1152/physiolgenomics.00214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Courtois-Coutry N, Le Moellic C, Boulkroun S, Fay M, Cluzeaud F, Escoubet B, Farman N, Blot-Chabaud M. Calcyclin is an early vasopressin-induced gene in the renal collecting duct. Role in the long term regulation of ion transport. J Biol Chem. 2002;277:25728–25734. doi: 10.1074/jbc.M112435200. [DOI] [PubMed] [Google Scholar]
- 42.Valenti G, Procino G, Tamma G, Carmosino M, Svelto M. Minireview: aquaporin 2 trafficking. Endocrinology. 2005;146:5063–5070. doi: 10.1210/en.2005-0868. [DOI] [PubMed] [Google Scholar]
- 43.Spiliotis ET, Nelson WJ. Here come the septins: novel polymers that coordinate intracellular functions and organization. J Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noda Y, Horikawa S, Furukawa T, Hirai K, Katayama Y, Asai T, Kuwahara M, Katagiri K, Kinashi T, Hattori M, Minato N, Sasaki S. Aquaporin-2 trafficking is regulated by PDZ-domain containing protein SPA-1. FEBS Lett. 2004;568:139–145. doi: 10.1016/j.febslet.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Stuart RO, Qiao J, Pavlova A, Bush KT, Pohl M, Sakurai H, Nigam SK. A role for Timeless in epithelial morphogenesis during kidney development. Proc Natl Acad Sci USA. 2000;97:10038–10043. doi: 10.1073/pnas.97.18.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.