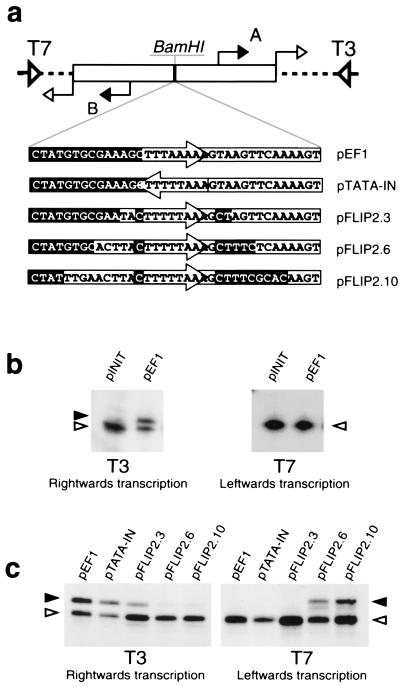
Figure 1.
Sequences flanking the TATA box are important for defining the orientation of transcription. (a) Diagram of plasmid constructs used in transcription assays. pINIT was created by ligating oligonucleotides corresponding to an inverted repeat of the T6 transcription initiation region into pBluescript. Subsequently, other oligonucleotides corresponding to derivatives of the Pyrococcus woesei ef1α promoter were inserted into the BamHI site of pINIT. The sequence of these inserts is shown. Sequences upstream of the TATA box in the wild-type promoter are shown in reverse shading; sequences naturally occurring downstream of the TATA box are boxed; and the TATA box itself is contained within an arrow. The T3 and T7 sequencing primer annealing sites are shown by open wide triangles and the T6 start sites are indicated by filled arrows and labeled A and B. The open arrows indicate the unexpected additional start sites arising at the junction of the cloned sequences and the parental polylinker that are recognized by the RNA polymerase alone and are totally independent of TBP and TFB. (b) The ef1α TATA box and flanking sequences direct unidirectional transcription. pINIT and pEF1 were used in in vitro transcription assays and transcripts detected by using primer extension with either T3 or T7 sequencing primer, as indicated. The transcript initiating at the downstream T6 start site is indicated with a solid arrowhead. The second, factor-independent, start site is indicated by an open arrowhead. The identities of the start sites were confirmed by electrophoresis adjacent to dideoxynucleotide DNA sequence ladders prepared with the radiolabeled T3 and T7 primers. (c) Sequences flanking the TATA box govern orientation of transcription. The various plasmid constructs shown in a were used in in vitro transcription assays. Annotation is as above.