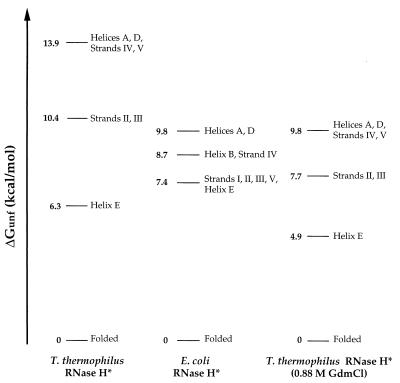
Figure 3.
Free energies of unfolding for partially and fully unfolded forms of T. thermophilus and E. coli RNases H*. Each line represents the difference in free energy between the native conformation and a form in which the structural elements indicated are unfolded; the form with the highest ΔGunf represents the fully unfolded conformation. Values of ΔGunf were averaged for each secondary structural unit, and regions were grouped together if the average of one region overlapped with the SD for another. Strand I is not represented for T. thermophilus RNase H*; one of its three tractable residues had a ΔGunf of 10.4 kcal/mol, whereas others were closer to the core region, both in their measured ΔGunfs and in their location in the structure. Stabilities in 0.88 M GdmCl were calculated for T. thermophilus RNase H* residues by using the parameters in Table 1, and values for coupled regions were averaged together.