Abstract
Background
Campylobacter is the most commonly reported bacterial cause of enteritis in humans in the EU Member States and other industrialized countries. One significant source of infection is broilers and consumption of undercooked broiler meat. Campylobacter jejuni is the Campylobacter sp. predominantly found in infected humans and colonized broilers. Sequence analysis of the 16S rRNA gene is very useful for identification of bacteria to genus and species level. The objectives in this study were to determine the degree of intraspecific variation in the 16S rRNA genes of C. jejuni and C. coli and to determine whether the 16S rRNA sequence types correlated with genotypes generated by PFGE analysis of SmaI restricted genomic DNA of the strains.
Methods
The 16S rRNA genes of 45 strains of C. jejuni and two C. coli strains isolated from broilers were sequenced and compared with 16S rRNA sequences retrieved from the Ribosomal Database Project or GenBank. The strains were also genotyped by PFGE after digestion with SmaI.
Results
Sequence analyses of the 16S rRNA genes revealed nine sequence types of the Campylobacter strains and the similarities between the different sequence types were in the range 99.6–99.9%. The number of nucleotide substitutions varied between one and six among the nine 16S rRNA sequence types. One of the nine 16S rRNA sequence profiles was common to 12 of the strains from our study and two of these were identified as Campylobacter coli by PCR/REA. The other 10 strains were identified as Campylobacter jejuni. Five of the nine sequence types were also found among the Campylobacter sequences deposited in GenBank. The three 16S rRNA genes in the analysed strains were identical within each individual strain for all 47 strains.
Conclusion
C. jejuni and C. coli seem to lack polymorphisms in their 16S rRNA gene, but phylogenetic analysis based on 16S rRNA sequences was not always sufficient for differentiation between C. jejuni and C. coli. The strains were grouped in two major clusters according to 16S rRNA, one cluster with only C. jejuni and the other with both C. jejuni and C. coli. Genotyping of the 47 strains by PFGE after digestion with SmaI resulted in 22 subtypes. A potential correlation was found between the SmaI profiles and the 16S rRNA sequences, as a certain SmaI type only appeared in one of the two major phylogenetic groups.
Background
Campylobacter spp., principally Campylobacter jejuni subsp.jejuni, are important food- and water-borne pathogens for man [1-3]. In the present paper Campylobacter jejuni subsp. jejuni is referred to as C. jejuni and Campylobacter jejuni subsp. doylei as C. doylei. Campylobacter jejuni is frequently found in the intestinal tract in a wide variety of wild and domesticated animals, especially poultry [1,4]. The genus Campylobacter was introduced by Sebald and Verón [5] and its taxonomic structure has been revised a number of times [6-8]. At present the genus Campylobacter contains 17 species, four of which have been further divided into eight subspecies [9]http://www.bacterio.cict.fr/. The use of molecular methods in bacteriology has resulted in increased knowledge about biodiversity within the genus Campylobacter. Sequence analysis of the 16S rRNA gene has proved extremely useful for evolutionary studies of prokaryotes [10] and constitutes the basis for the revised taxonomy of bacteria [11]. This method has also been successfully used for identification of Campylobacter spp. [12]. Taxonomy has become a flexible and constantly evolving science and further progress in methodologies will probably result in additional adjustments in the classification of members of the genus Campylobacter. Furthermore, several PCR assays have successfully been applied for detection of Campylobacter with improved accuracy in identification of Campylobacter spp. from various sources.
Isolates from different bacteria can be identified to genus and often also to species level by sequence analysis of the 16S rRNA gene. Most species within the genus Campylobacter can be successfully differentiated by sequence analysis of the 16S rRNA gene. However, C. jejuni, C. coli and C. lari have been more difficult to differentiate by such sequence analysis [12]. Bacterial genomes can harbour up to 15 rRNA operons and the sequences of the corresponding genes are not always identical, which can make it difficult to interpret sequence data [13]. The nucleotide substitutions between different genes within a gene family are referred to as polymorphisms. Sequence differences can also occur between homologous genes from different strains of a certain bacterial species. All these nucleotide substitutions are collectively referred to as intraspecific variation. It has previously been reported that C. jejuni has three copies of the 16S rRNA gene in the genome [14,15]. One aim of our study was to establish if sequence differences exist between the three genes in a certain Campylobacter jejuni strain, because if intraspecific variation exists in these genes it may be possible to use the method for subtyping [16,17]. In a previous study of Campylobacter spp. from broilers in Sweden, 390 isolates were analysed by PFGE [18]. After digestion with SmaI, almost 80 different banding patterns (SmaI types) were obtained, including isolates that could not be digested with SmaI. Some of the subtypes were isolated more frequently than others. Another aim of the present study was to determine the sequence variation in the 16S rRNA genes of different Campylobacter jejuni strains and to determine whether the 16S rRNA sequence types correlated to the SmaI types.
Materials and methods
Bacterial strains
All bacterial strains were isolated from cloacal samples collected at slaughter within the Swedish Campylobacter programmes. The origins of the 47 strains (producer and slaughterhouse) are given in Table 1. The strains were isolated from different flocks delivered by 21 broiler producers to 6 different slaughterhouses during two periods, 1995–1997 and 2002–2004. First, two strains from each of the 10 most common SmaI types reported within the Campylobacter program in Sweden 2002–2004 were selected. Then eight new strains found at least four times in a previous Swedish study [18] were also selected for analysis. The selection was based on the SmaI profiles, and the profiles of these strains were similar to those of the 10 most common SmaI types. The 19 strains from 1995–1997 (Table 1) were chosen from the same producers as those from 2002–2004, which are known to frequently deliver Campylobacter-positive broilers [19]. The SmaI profiles are numbered according to previous studies in Sweden [18].
Table 1.
Forty seven strains of C. jejuni and C. coli from cloacal samples taken within the Swedish Campylobacter programmes for broilers, for which the 16S rRNA gene sequences were determined in the present study
| Isolate | 16S rRNA seq. type | PFGE SmaI1 | Year | Acc number2 | Producer3 | Slaughter- house3 |
| 13860/02 | A | 3 | 2002 | EU127502 | 4 | I |
| 8266/02 | A | 3 | 2002 | EU127503 | 7 | II |
| 13168/02 | A | 2 | 2002 | EU127504 | 3 | VI |
| 3105-2/96 | A | 2 | 1996 | EU127505 | 6 | V |
| 17903/02 | B | 4 | 2002 | EU127506 | 20 | III |
| 18381/02 | B | 4 | 2002 | EU127507 | 16 | II |
| 12739/02 | B | 10 | 2002 | EU127508 | 10 | V |
| 10754/03 | B | 10 | 2003 | EU127509 | 5 | VI |
| 1132-3/95 | B | 2 | 1995 | EU127510 | 18 | II |
| 5169-1/96 | B | 2 | 1996 | EU127511 | 4 | I |
| 5169-2/96 | B | 2 | 1996 | EU127512 | 4 | I |
| 5174-2/96 | B | 2 | 1996 | EU127513 | 4 | I |
| 6133-1/96 | B | 2 | 1996 | EU127514 | 8 | III |
| 6188-1/96 | B | 2 | 1996 | EU127515 | 7 | III |
| 11532/04 | B | 36 | 2004 | EU127516 | 18 | II |
| 9612/04 | B | 42 | 2004 | EU127517 | 10 | V |
| 10051/03 | B | 2 | 2003 | EU127518 | 11 | V |
| 6113/97 | B | 2 | 1997 | EU127519 | 20 | III |
| 6114/97 | B | 4 | 1997 | EU127520 | 20 | III |
| 11036/96 | C | 2 | 1996 | EU127521 | 19 | IV |
| 1160-2/96 | D | 10 | 1996 | EU127522 | 16 | II |
| 1160-3/96 | D | 10 | 1996 | EU127523 | 16 | II |
| 10056/03 | E | 5 | 2003 | EU127524 | 15 | II |
| 10942/03 | E | 6 | 2003 | EU127525 | 16 | II |
| 10713/03 | E | 8 | 2003 | EU127526 | 14 | V |
| 11414/03 | E | 8 | 2003 | EU127527 | 1 | II |
| 12717/02 | E | 9 | 2002 | EU127528 | 2 | III |
| 3243/02 | E | 9 | 2002 | EU127529 | 8 | III |
| 8693/04 | E | 67 | 2004 | EU127530 | 18 | II |
| 11318/04 | E | 70 | 2004 | EU127531 | 6 | V |
| 1167-3/95 | E | 101 | 1995 | EU127532 | 18 | II |
| 1182-3/95 | E | 102 | 1995 | EU127533 | 9 | II |
| 1184-3/95 | E | 102 | 1995 | EU127534 | 9 | II |
| 8103/04 | E | 57 | 2004 | EU127535 | 12 | V |
| 9582/04 | F | 26 | 2004 | EU127536 | 19 | V |
| 10626/04 | G | 15 | 2004 | EU127537 | 9 | II |
| 19280/02 | H | 1 | 2002 | EU127538 | 21 | I |
| 18279/02 | H | 1 | 2004 | EU127539 | 4 | I |
| 8525/02 | H | 5 | 2002 | EU127540 | 17 | I |
| 12279/02 | H | 7 | 2002 | EU127541 | 12 | I |
| 15193/03 | H | 7 | 2003 | EU127542 | 8 | III |
| 5304/04 | H | 16 | 2004 | EU127543 | 9 | II |
| 11020/96 | H | 20 | 1996 | EU127544 | 19 | IV |
| 6144/96 | H | 5 | 1996 | EU127545 | 2 | III |
| 6181/96 | H | 6 | 1996 | EU127546 | 2 | III |
| 9916/03 | H | 6 | 2003 | EU127547 | 17 | I |
| 11281/04 | I | 56 | 2004 | EU127548 | 6 | V |
1 The numbering of SmaI types 1 to 100 is according to other studies [15]. SmaI types assigned numbers greater than 100 have not been found previously in subtyping of isolates within the Swedish Campylobacter program 2001–2006.
2 Accession number in GenBank.
3 Origin of the isolates.
Procedures for sampling, transport and laboratory analyses have been described previously [19]. Identification of Campylobacter spp. was based on colony morphology, microscopic appearance and the following phenotypic characteristics: motility, production of cytochrome C oxidase and catalase, and the hippurate hydrolysis reaction [20]. One colony from each positive sample was stored in glycerol broth (15% glycerol and 85% serum broth) at -70°C before further use.
Genotyping of selected strains by PCR/REA and PFGE
Species identification of C. jejuni was initially based on a positive hippurate hydrolysis reaction. Isolates that gave a weak or negative hippurate reaction were speciated by the polymerase chain reaction, followed by restriction enzyme analysis (PCR/REA) by which C. jejuni, C. coli, C. lari, and C. upsaliensis can be identified and differentiated [21]. In addition, some strains were tested by PCR for the hippuricase gene [22].
Genetic subtyping of C. jejuni isolates was performed by pulsed field gel electrophoresis (PFGE) in accordance with the standardised Campynet procedure [23]. Genomic DNA was digested with SmaI and the fragments were separated by PFGE in a CHEF-DRII apparatus (BioRad Laboratories, Hercules, CA, USA). The PFGE banding patterns were analysed by computer-assisted identification with GelCompar II (Applied Maths, Kortrijk, Belgium).
Sequencing of 16S rRNA genes
DNA was prepared from the bacteria by lysis of 1–3 colonies in 100 μl of water at 100°C for 10 min. The lysate was used as a template for in vitro amplification of the 16S rRNA genes by PCR. The PCR products were used for cycle sequencing with fluorescently labelled terminators (Big Dye; Applied Biosystems, Foster City, Calif., USA) as described by the manufacturer and with a set of sequencing primers developed for members of the phylum Proteobacteria [24]. The sequencing products were analysed by capillary electrophoresis on an ABI Prism 3100 genetic analyser (Applied Biosystems). Contiguous sequences (contigs) were generated by using the program Contig Express included in the Vector NTI Suite (InforMax, Bethesda, Md., USA). The contigs were checked and edited manually if necessary before phylogenetic analysis and deposition in GenBank. The accession numbers for the sequences in GenBank are given in Table 1.
Phylogenetic analysis
The 16S rRNA sequences determined in this work were aligned manually with prealigned sequences retrieved from the Ribosomal Database Project II [25] and by using the Genetic Data Environment software [26]. The phylogenetic trees were constructed by neighbour-joining [27] from a distance matrix that was corrected for multiple substitutions at single locations by the two-parameter method [28]. The distance matrix comprised 1417 nucleotide positions corresponding to positions 36 to 1472 in the 16S rRNA sequence of Escherichia coli.
Results
Identification of the isolates
Forty-five of the 47 strains included in this study were identified as Campylobacter jejuni by a positive hippurate hydrolysis reaction or by PCR/REA or by the presence of the hippuricase gene as judged from PCR. Two strains were identified as C. coli (8693/04 and 11318/04) by the same methods. All isolates were identified as representing either C. jejuni or C. coli by 16S rRNA sequence analysis.
Sequence analysis of the 16S rRNA genes
The length of the 16S rRNA gene fragments of the sequenced Campylobacter isolates was 1417 nucleotides. Ambiguities were not found in any of the sequences, which showed that there were no sequence polymorphisms in the three 16S rRNA genes. However, nucleotide substitutions were found in eight positions of the strains sequenced in the present study, which resulted in nine different 16S rRNA sequence types, referred to as a-i in Tables 1, 2 and 3. The nucleotide sequences of the 16S rRNA genes of the 47 strains were determined and compared with 16S rRNA sequences of 21 strains of C. jejuni, 2 strains of C. doylei, one strain each of C. lari and C. upsaliensis and 16 strains of C. coli retrieved from RDP-II (Table 2). Nine additional positions that were variable were identified in the sequences deposited by other authors (Table 3). The Swedish strains were not variable in these positions. The sequence similarities between the nine different sequence types (a-i) were in the range 99.6–99.9%. The sequence similarities among C. jejuni strains and among C. coli strains were in the range 99.5–99.9% and 98.2–99.9%, respectively. Whole genome sequences are available for four of the C. jejuni strains (accession numbers, CP000025, AL111168, CP000814 and CP000538). Whole genome sequence data show that the genomes of Campylobacter jejuni strains harbour three rRNA operons and we found that the sequences of the three 16S rRNA genes were identical within each individual strain for all 47 strains. Furthermore, the three 16S rRNA genes of C. doylei (CP000768) are also identical. The nine profiles occurred with varying frequencies among the 47 strains analysed here. Three 16S rRNA sequence types (b, e and h) dominated and constituted 79% (37 of 47 strains). The number of nucleotide substitutions varied between one and six among the nine 16S rRNA sequence types. The largest difference was found between the 16S rRNA sequence types f and i (see Fig. 1). Furthermore, five of the sequence types (b, d, e, h and i) had identical 16S rRNA sequence profiles to strains retrieved from GenBank. Sequence type (e) was shared between 12 of the strains in our study and two of these were identified as C. coli by PCR/REA. These strains had an identical 16S rRNA profile to C. jejuni (CP000025), for which the whole genome sequence is available, and also to one strain of C. coli (AF550623).
Table 2.
Campylobacter spp. strains for which the 16S rRNA sequences were retrieved from RDP-II or GenBank and used in the phylogenetic analysis
| Species | Strain | Acc no in GenBank | Acc no to an identical seq | No of Ns4 in the seq | Identical to Seq type |
| C. coli | LMG 6440 | AF372092 | |||
| C. coli | LMG 9220 | AF550620 | |||
| C. coli | LMG 15883 | AF550621 | |||
| C. coli | LMG 15884 | AF550622 | |||
| C. coli | H99/155 | AF550624 | |||
| C. coli | B99/131 | AF550625 | |||
| C. coli | ATCC 49941 | AY621115 | |||
| C. coli | NZ1905-94 | DQ174136 | |||
| C. coli | NZ2695-96 | DQ174137 | |||
| C. coli | CCUG 11283 | L04312 | 10 | ||
| C. coli | RMIT32A | L19738 | 12 | ||
| C. coli | H99/119 | AF550623 | e | ||
| C. coli | Lio8 | DQ1741353 | AF550623 | e | |
| C. coli | NZ899-00 | DQ1741383 | AF550623 | e | |
| C. coli | NZ900-95 | DQ1741393 | AF550623 | e | |
| C. coli | NZ4812-94 | DQ1741403 | AF550623 | e | |
| C. jejuni | RM12212 | CP000025 | e | ||
| C. jejuni | 98/E600/5 | AF3932023 | RM1221 | e | |
| C. jejuni | 98/E599/10 | AF3932033 | RM1221 | e | |
| C. jejuni | LMG9217 | AF5506263 | RM1221 | e | |
| C. jejuni | CCUG 10937 | DQ1741413 | RM1221 | e | |
| C. jejuni | NCTC 11351 | AF372091 | |||
| C. jejuni | CCUG 11284 | L04315 | 9 | ||
| C. jejuni | WH11 | AF3932043 | L04315 | d | |
| C. jejuni | BB/224 | AF5506293 | L04315 | d | |
| C. jejuni | LMG 9243 | AF550627 | |||
| C. jejuni | H99/240 | AF550628 | |||
| C. jejuni | TGH9011 | Z29326 | |||
| C. jejuni | SSI 5384-98 | Y19244 | |||
| C. jejuni | NCTC 111681,2 | AL111168 | |||
| C. jejuni | 6871 | AY628389 | |||
| C. jejuni | 81–1762 | CP000538 | i | ||
| C. jejuni | ATCC 294228 | DQ174142 | h | ||
| C. jejuni | B99/206 | AF5506303 | DQ174142 | h | |
| C. jejuni | Lio6 | DQ174143 | b | ||
| C. jejuni | ATCC 49943 | AY6211123 | DQ174143 | b | |
| C. jejuni | CCUG 24567 | L146303 | DQ174144 | ||
| C. doylei | LMG8843 | DQ174144 | |||
| C. doylei | Not defined | AY6211113 | DQ174143 | ||
| C. lari | Not defined | L04316 | 6 | ||
| C. upsaliensis | Not defined | L14628 | 11 |
1 Strain for which the whole genome sequence is available.
2 The sequence without deletions was used in the tree.
3 This sequence was not used in the tree (Fig. 2), because it was identical to another sequence.
4 Number of ambiguities in the sequence, which also reflects the sequencing accuracy.
Table 3.
Polymorphic positions in the nine different 16S rRNA sequence types obtained from C. jejuni isolated from cloacal samples taken within the Swedish Campylobacter program for broilers.
| Seq type | No of isolates | 80 | 126 | 141* | 205 | 554* | 614* | 644 | 687* | 703* | 712* | 814 | 821* | 986 | 995 | 1228* | 1244 | 1400* |
| A | 4 | C | A | C | C | C | G | C | C | G | C | A | T | A | T | C | C | T |
| B | 15 | C | A | C | T | C | G | C | C | G | C | A | T | A | T | C | C | T |
| C | 1 | C | A | C | T | C | G | C | C | G | C | A | T | A | T | C | T | T |
| D | 2 | C | A | C | T | C | G | C | C | G | C | G | T | A | T | C | C | T |
| E | 12 | C | A | C | T | C | G | C | C | G | C | G | T | T | A | C | C | T |
| F | 1 | C | G | C | T | C | G | C | C | G | C | G | T | T | A | C | C | T |
| G | 1 | T | A | C | T | C | G | C | C | G | C | A | T | A | T | C | C | T |
| H | 10 | T | A | C | T | C | G | C | C | G | C | G | T | T | A | C | C | T |
| i | 1 | T | A | C | T | C | G | T | C | G | C | A | T | A | T | C | C | T |
| CP000025 | 1 | C | A | C | T | C | G | C | C | G | C | G | T | T | A | C | C | T |
| AL111168 | 1 | T | A | C | T | C | G | C | C | G | C | G | T | A | T | C | C | T |
*Positions where some of the C. jejuni 16S rRNA sequences deposited in GenBank by other authors differed from the sequences determined in this study
The nucleotide positions were numbered according to one of the 16S rRNA gene sequences of C. jejuni strain NCTC 11168, for which the whole genome sequence is available from GenBank. However, the 16S rRNA sequence was retrieved from RDP-II [29].
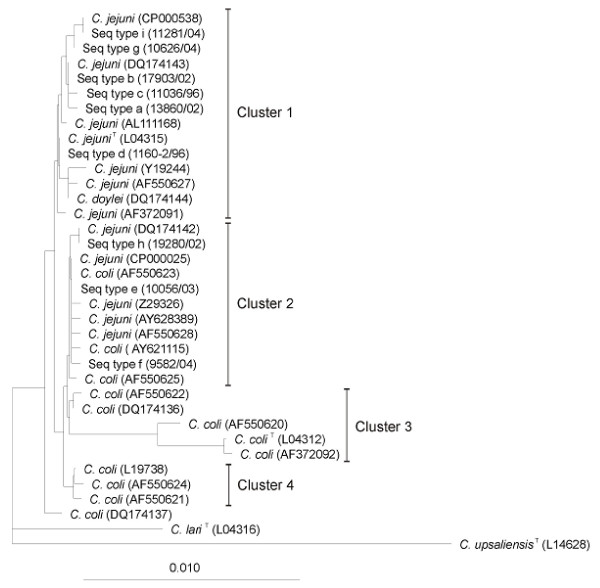
Figure 1.
Radial representation of a phylogenetic tree constructed by the neighbour-joining method showing the relationships between the different sequence types (Seq type), C. coli and C. jejuni. Campylobacter coli was used as outgroup. The length of the scale bar represents one nucleotide substitution in the 16S rRNA gene fragment (1417 nucleotides).
Phylogenetic analysis
The strains in the present study were grouped into two major clusters in the radial tree (Fig. 1). The first cluster comprised sequence types a, b, c, d, g and i and contained sequence types of C. jejuni retrieved from GenBank (Fig. 2). The sequence types a, b, c, g and i had the nucleotides A, A and T in the positions 814, 986 and 995, respectively, whereas sequence type d had a G in the first of these positions (Table 3). The SmaI types in this group were 2, 3, 4, 10, 15, 36, 42 and 56. The second cluster comprised sequence types e, f and h and contained both C. coli and C. jejuni. These strains are characterised by having G, T, and A in nucleotide positions 814, 986 and 995, respectively (Table 3). The corresponding SmaI types in this group were 1, 5, 6, 7, 8, 9, 16, 20, 26, 57, 67, 70, 101 and 102.
Figure 2.
Evolutionary tree showing the phylogenetic relationships between C. lari, C. doylei, C. coli and C. jejuni retrieved from RDP-II and the nine sequence types (Seq type) of Campylobacter spp. identified in this study. A representative strain is given in brackets after the sequence types. Accession numbers in GenBank are given for the other strains. Camplyobacter upsaliensis was used as outgroup. The length of the scale bar represents 1 nucleotide substitution per 100 positions.
Phylogenetic relationships of the nine 16S rRNA sequence types obtained in the present study and certain strains of C. jejuni, C. coli, C. doylei, C. lari and C. upsaliensis reported by other authors are shown in Fig. 2. The taxa in this tree form four clusters (1–4) and one single species line (C. coli, DQ174137). However, due to the high-sequence similarity, the nodes are not very stable and sequencing errors in some of the older sequences may well affect the topology of the tree. Cluster 1 contains only C. jejuni and C. doylei strains. Cluster 2 contains both C. jejuni and C. coli strains, while clusters 3 and 4 contain only C. coli strains. The three C. coli strains (AF550620, L04312 and AF372092) have sequence similarities to C. jejuni in the range 98.3–98.6%.
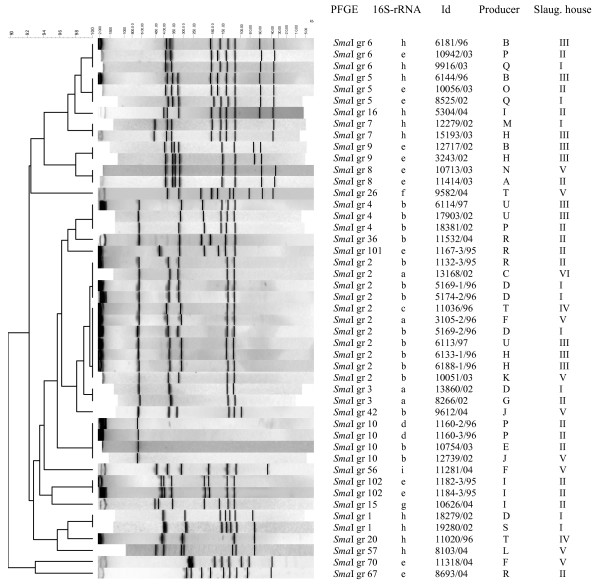
Pulsed-field gel electrophoresis
The 47 strains were subtyped into 22 different SmaI types, including the strains refractory to SmaI digestion (designated SmaI type 10). Of the 22 SmaI types, 11 were found only once, and the remaining 11 subtypes were found at least twice (Fig. 3). The subtype that was most often observed was SmaI type 2 (11 isolates) and this was also a subtype that was widely spread among producers, slaughterhouses and years. No correlation could be found between the different 16S rRNA sequences or SmaI types and the year, slaughterhouse or farm from which the Campylobacter strain had been isolated. Strains with the same SmaI type but of different sequence types appeared only in one the two phylogenetic clusters (Fig 1).
Figure 3.
Dendrogram of the PFGE profiles obtained after SmaI digestion of DNA from Campylobacter jejuni and C. coli strains from cloacal samples taken within the Swedish Campylobacter program for broilers. The numbering of SmaI types 1 to 100 is according to other studies [15]. SmaI types assigned numbers greater than 100 have not been found previously in subtyping of isolates within the Swedish Campylobacter program.
Discussion
Genomes of C. jejuni and C. coli only harbour three copies of the 16S rRNA genes, whereas E. coli has seven rRNA genomic loci [14,29,30]. The three 16S rRNA gene sequences of the respective 47 strains included in the study were identical as judged from the sequence raw data, which would have revealed any polymorphism. This finding was surprising because bacteria with more than one rRNA operon often have at least a few polymorphisms in their 16S rRNA genes. The observation that C. jejuni seem to lack polymorphisms in their 16S rRNA genes is also supported by genome sequencing data, because three of the strains (CP000025, CP000814 and AL111168) have identical 16S rRNA genes. The third strain (CP000538) has two deletions in one of the three genes, but except for that, the three genes are also identical. We did not find sequence length polymorphisms and in this respect C. jejuni is very stable. Reparation mechanisms for concerted evolution of rRNA genes seem to be very efficient. Furthermore, the whole genome sequence of Campylobacter doylei (CP000768) also contains three identical 16S rRNA sequences.
It was possible to identify eight signature nucleotide positions by which the studied strains of C. jejuni could be differentiated into nine groups according to their 16S rRNA sequences. The observed nucleotide substitutions at different positions in the 16S rRNA genes of Campylobacter spp. were compared with the positions of the nucleotide substitutions that appear as mutations in E. coli [31]. However, none of the nucleotide substitutions was found at the homologous positions. Restriction analysis by SmaI digestion and PFGE was, however, a more discriminatory typing method, detecting 22 subtypes as compared with nine different 16S rRNA sequence types. The discriminatory power of PFGE for genotyping of Campylobacter spp. has been reported previously [32,33]. One sequence type (e) had identical 16S rRNA sequence as one of the whole genome sequenced C. jejuni strains (RM1221) and C. coli strain (Lio8), (Table 2, Fig 2). Furthermore, sequence type e differed in only two nucleotide positions from C. coli (L19738). Two of the isolates (8693/04 and 11318/04) belonging to16S rRNA type e were also identified as C. coli by PCR/REA. This observation shows that phylogenetic analysis based on 16S rRNA cannot always be used to differentiate between C. jejuni and C. coli, although in cluster 3 (Fig. 2), three of the C. coli sequences are significantly different from the C. jejuni sequences. However, the results must be interpreted with care, because sequences of many mistyped bacterial strains have been deposited in GenBank. The results in the present study confirm the limitations of the 16S rRNA gene for resolving close, relationships.
The common SmaI types identified in Swedish broilers in 2002–2004 were also found among isolates from 1995–1997, a fact that indicates that these common types are also stable over time. Furthermore, the whole genome sequenced C. jejuni (CP000025) with an identical profile to sequence type e originates from chickens in the USA [34]. This raises the question whether certain clones are more common than others and whether those clones have a higher ability to survive in the environment around the broilers or within animals around the broiler houses over a very long time. Bacteria with relatively small genomes, such as C. jejuni, may undergo genetic variation to increase their potential to adapt to new environments [14]. Such genotypic variation could result in phenotypic changes. These variations are probably important in the transmission route from broiler to man, where Campylobacter spp. must survive several hostile environments.
Conclusion
Bacterial genomes could harbour up to fifteen 16S rRNA operons. Polymorphisms in the 16S rRNA gene are common in bacteria with more than one 16S rRNA operon. Genomes of Campylobacter spp. harbour three copies of the 16S rRNA genes. Polymorphisms were not found in the 16S rRNA genes from any of the 47 Campylobacter spp. strains sequenced. The three rRNA operons in the analysed Campylobacter jejuni strains were identical within each individual strain for all 47 strains, which indicates that C. jejuni and C. coli are very stable in this respect.
Genotyping of 47 strains by 16S rRNA gene sequencing resulted in nine sequence types, whereas PFGE after digestion with SmaI resulted in 22 subtypes. Phylogenetic analysis based on 16S rRNA sequences is not always sufficient for differentiation between C. jejuni and C. coli. A potential correlation was found between the SmaI profiles and the 16S rRNA sequences, as a certain SmaI type only appeared in one of the two major phylogenetic groups.
Authors' contributions
IH, EOE and KEJ participated in the discussion on the study design, the collection of isolates, analysis and interpretation of data, and in the writing of the manuscript, EOE and LS carried out the analysis and interpretation of PFGE data. Analysis and interpretation of sequence data of the 16S rRNA gene were carried out by IH, MP and KEJ. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors gratefully acknowledge the staff at the Department of Bacteriology, National Veterinary Institute, and the Department of Biomedical Sciences and Veterinary Public Health, Swedish University of Agricultural Sciences, for assistance with the microbiological analyses and the staff at the slaughterhouses participating in the study and thus making this work possible. This work was supported by the Ivar and Elsa Sandberg Research Foundation and the Swedish Farmers' Foundation for Agricultural Research.
Contributor Information
Ingrid Hansson, Email: ingrid.hansson@sva.se.
Marianne Persson, Email: marianne.persson@sva.se.
Linda Svensson, Email: linda.svensson@sva.se.
Eva Olsson Engvall, Email: eva.olsson@sva.se.
Karl-Erik Johansson, Email: karl-erik.johansson@bvf.slu.se.
References
- Skirrow MB. Diseases due to Campylobacter, Helicobacter and related bacteria. J Comp Pathol. 1994;111(2):113–149. doi: 10.1016/S0021-9975(05)80046-5. [DOI] [PubMed] [Google Scholar]
- Bryan FL, Doyle MP. Health risks and consequences of Salmonella and Campylobacter jejuni in raw poultry. J Food Prot. 1995;58:326–344. doi: 10.4315/0362-028X-58.3.326. [DOI] [PubMed] [Google Scholar]
- Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. Campylobacter jejuni – an emerging foodborne pathogen. Emerg Infect Dis. 1999;5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Taylor DN, Feldman RA. Epidemiology of Campylobacter jejuni infections. Epidemiol Rev. 1983;5:157–173. doi: 10.1093/oxfordjournals.epirev.a036256. [DOI] [PubMed] [Google Scholar]
- Sebald ER, Véron M. DNA base content and classification of vibrios (In French; Teneur en bases da lÀDN et classification des Vibrions) Annales de L'institut Pasteur (Paris) 1963;105:897–910. [PubMed] [Google Scholar]
- Goodwin CS, Armstrong JA, Chilvers T, Peters M, Collins MD, Sly L, McConnell W, Harper WES. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov., respectively. J Syst Bact. 1989;39:397–405. [Google Scholar]
- Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: Emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- Vandamme P, On SLW. Recommendations of the subcommittee on the taxonomy of Campylobacter and related bacteria. Int J Syst Evol Microbiol. 2001;51:719–721. doi: 10.1099/00207713-51-2-719. [DOI] [PubMed] [Google Scholar]
- List of procaryotic names with standing in Nomenclature http://www.bacterio.cict.fr/
- Woese CR. Bacterial evolution. Microbiol Reviews. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Klenk HP. A phylogenetic backbone and taxonomic framework for procaryotic systematics. In: Boone DR, Castenholz RW, editor. Bergey's Manual of Systematic Bacteriology The Archaea and the Deeply Branching and Phototrophic Bacteria. Vol. 1. Berlin, Springer; 2001. pp. 49–65. [Google Scholar]
- Gorkiewicz G, Feierl G, Schober C, Dieber F, Köfer J, Zechner R, Zechner EL. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J Clin Microbiol. 2003;41:2537–2546. doi: 10.1128/JCM.41.6.2537-2546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Taylor DE, Eaton M, Yan W, Chang N. Genome maps of Campylobacter jejuni and C. coli. J Bacteriol. 1992;174:2332–2337. doi: 10.1128/jb.174.7.2332-2337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Lombardi R, Bingham H, Hani E, Louie H, Ng D, Chan VL. Fine mapping of the three rRNA operons on the updated genomic map of Campylobacter jejuni TGH9011 (ATCC 43431) J Bacteriol. 1993;175:7468–7470. doi: 10.1128/jb.175.22.7468-7470.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldtander Königsson M, Bölske G, Johansson KE. Intraspecific variation in the 16S rRNA gene sequences of Mycoplasma agalactiae and Mycoplasma bovis strains. Vet Microbiol. 2002;85:209–220. doi: 10.1016/S0378-1135(01)00517-X. [DOI] [PubMed] [Google Scholar]
- Heldtander M, Wesonga H, Bölske G, Pettersson B, Johansson KE. Genetic diversity and evolution of Mycoplasma capricolum subsp. capripneumoniae strains from eastern Africa assessed by 16S rDNA sequence analysis. Vet Microbiol. 2001;78:13–28. doi: 10.1016/S0378-1135(00)00290-X. [DOI] [PubMed] [Google Scholar]
- Hansson I, Vågsholm I, Svensson L, Engvall EO. Correlations between Campylobacter spp. prevalence in the environment and broiler flocks. J Appl Microbiol. 2007;103:640–649. doi: 10.1111/j.1365-2672.2007.03291.x. [DOI] [PubMed] [Google Scholar]
- Hansson I, Engvall EO, Lindblad J, Gunnarson A, Vågsholm I. The Campylobacter surveillance program for broilers in Sweden, July 2001–June 2002. Vet Rec. 2004;155:193–196. doi: 10.1136/vr.155.7.193. [DOI] [PubMed] [Google Scholar]
- Nachamkin I. Campylobacter and Arcobacter. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editor. Manual of Clinical Microbiology. ASM Press, Washington, DC; 1995. pp. 113–117. [Google Scholar]
- Fermér C, Engvall EO. Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J Clin Microbiol. 1999;37:3370–3373. doi: 10.1128/jcm.37.10.3370-3373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D, Lawson AJ, Owen RJ, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standardised Campynet procedure http://www.svs.dk/campynet/PFGE.html
- Båverud V, Nyström C, Johansson KE. Isolation and identification of Taylorella asinigenitalis from the genital tract of a stallion, first case of a natural infection. Vet Microbiol. 2006;116:294–300. doi: 10.1016/j.vetmic.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33(Database issue):D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. Version 22 Millipore Imaging Systems. Ann Arbor, MI, USA; 1992. GDE: Genetic Data Environment. [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Nuijten PJM, Bartels C, Bleumink-Pluym C, Gaastra W, Zeijst BAM van der. Size and physical map of the Campylobacter jejuni chromosome. Nucleic Acids Res. 1990;18:6211–6214. doi: 10.1093/nar/18.21.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Bingham H, Khawaja R, Louie H, Hani E, Neote K, Chan VL. Physical map of Campylobacter jejuni TGH9011 and localisation of 10 genetic markers by use of pulsed-field gel electrophoresis. J Bacteriol. 1992;174:3494–3498. doi: 10.1128/jb.174.11.3494-3498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triman KL. The 16S ribosomal RNA mutation database (16SMDB) Nucleic Acids Res. 1996;24:166–168. doi: 10.1093/nar/24.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T, Geilhausen B, Newell D. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Duim B, Rigter A, Plas J van der, Jacobs-Reitsma WF, Wagenaar JA. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000;38:1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Quinones B, Miller WG, Horn ST, Mandrell RE. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J Clin Microbiol. 2006;44:4125–4135. doi: 10.1128/JCM.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]