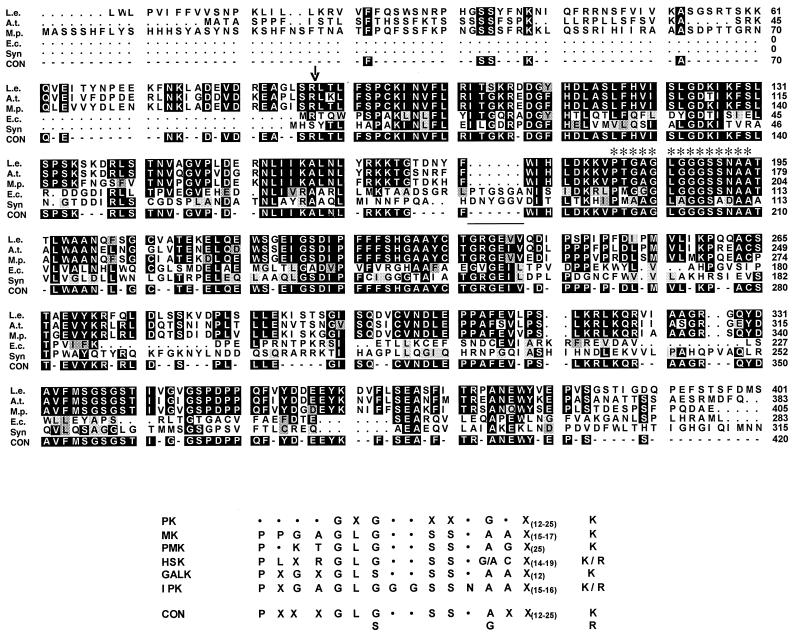
Figure 3.
(Upper) Deduced amino acid sequence comparison of IPK from peppermint (M.p.; AF179283), pTOM41 from tomato (L.e.; U62773), a hypothetical protein from A. thaliana (A.t.; AC005168, PID g3426035), IPK from E. coli (E.c.; AF179284), and a hypothetical protein from Synechocystis sp. strain PC6803 (Syn; D90899, PID g16665). CON indicates the consensus sequence. Identical residues are black with white lettering, residues of high similarity are indicated in gray, and residues of lower similarity are in light gray. Asterisks indicate the position of the putative ATP-binding motif, the putative cleavage site of the plastidial targeting sequence is shown by an arrow, and a conserved insert in bacterial sequences is underlined. (Lower) Comparison of sequences of an ATP-binding site found in representatives of protein kinases (PK), mevalonate kinases (MK), phosphomevalonate kinases (PMK), homoserine kinases (HSK), galactokinases (GALK), and isopentenyl monophosphate kinases (IPK).