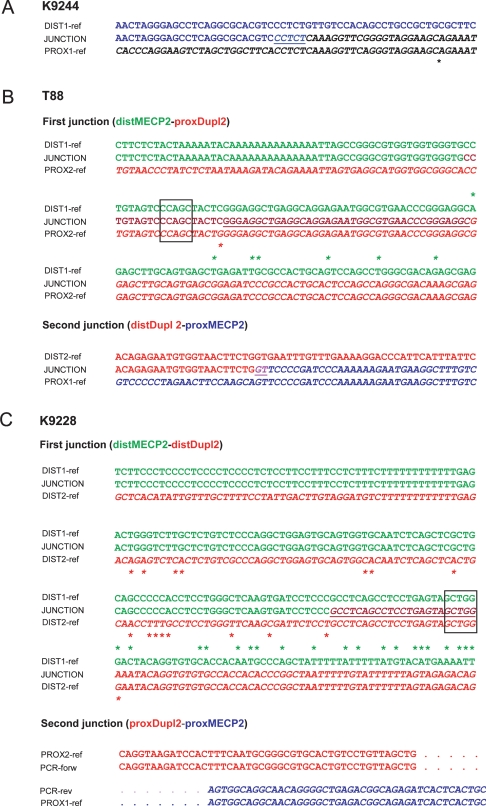
Figure 4.
Alignment of the sequenced junctions in patients K9244, T88, and K9228 with the reference genome sequence. Proximal and distal reference sequences are printed in normal font and italics, respectively, and colored differently. All junctions are underlined and in the merge color of proximal and distal reference sequence. Asterisks represent sequence mismatches. (A) K9244: Proximal (top) and distal (bottom) sequence were aligned against the junction sequence (middle), demonstrating the tandem orientation of the duplication. A microhomology of 5 bp (CCTCT) is found at the junction between the distal and proximal sequences, characteristic for NHEJ. (B) T88: Proximal (top) and distal (bottom) sequence of the first and second junction were aligned against the respective junction sequences (middle). The first junction occurs within an AluY present at both dBkpnt and pBkpnt of the MECP2 and the distal duplication, respectively. There is a perfect match of 39 bp. The Alu pentanucleotide motif (CCAGC) is boxed. These data are consistent with homology-assisted NHEJ. A microhomology of 2 bp (GT) is found at the second junction, in agreement with a collapse of the BIR-fork at this locus and search for microhomology on the sister chromatid. (C) K9228: Alignment of the reference sequence against the respective junction sequences (middle). The first junction occurs within an AluJo at the dBkpnt of the MECP2 duplication and an AluSg at the dBkpnt of the distal duplication. The homology-assisted NHEJ at this junction results in the insertion of the distal duplication in between the MECP2 duplications in an inverted orientation. The 23-bp junction terminates in the Alu pentanucleotide motif (boxed). Boundaries of the PCR-fragment containing the second junction were sequenced and revealed sequences of the pBkpnt of the MECP2 duplication at one side (PCR-forw) while at the other end (PCR-rev) sequences of LCR DC1649 were obtained, coinciding with the pBkpnt of the distal duplication. Dots represent predicted reference sequence.