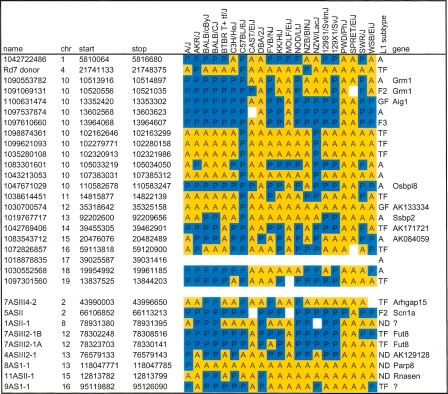
Table 2.
Validation of L1 polymorphisms in classical and wild mouse strains
(Top) Candidate L1 polymorphisms from chromosomes 10 and others were arbitrarily selected as described in the text for validation by PCR. (Bottom) Nine putative L1 integrants from a screen of fusion transcripts were identified in unassembled strains by chromosome walking. PCR reactions across left and right genomic junctions and empty target sites validated presence (blue, P) or absence (yellow, A) of individual integrants, as predicted by WGS trace alignments for four unassembled strains. In a few cases, no PCR product was obtained (white), suggesting additional genetic variation in a strain, or suboptimal PCR design. Trace ID or cDNA clone names, chromosomal coordinates, spanning gene names, and L1 subtypes from RepeatMasker classification and Cross_Match reclassification are indicated (see Supplemental Methods).