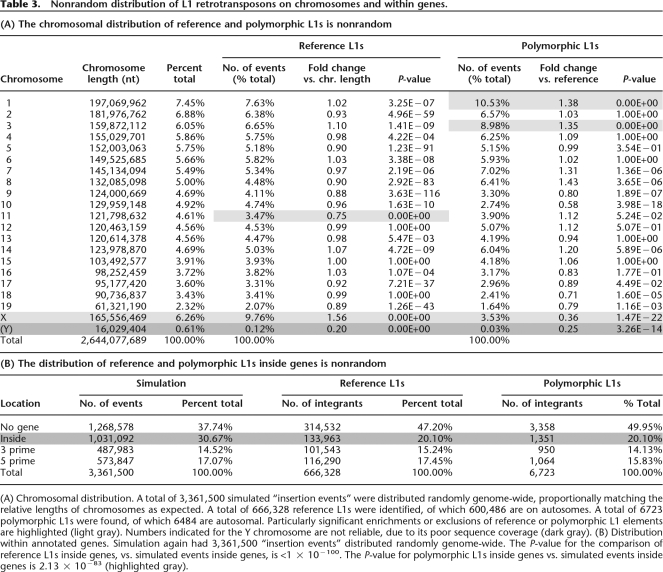
Table 3.
Nonrandom distribution of L1 retrotransposons on chromosomes and within genes.
(A) Chromosomal distribution. A total of 3,361,500 simulated “insertion events” were distributed randomly genome-wide, proportionally matching the relative lengths of chromosomes as expected. A total of 666,328 reference L1s were identified, of which 600,486 are on autosomes. A total of 6723 polymorphic L1s were found, of which 6484 are autosomal. Particularly significant enrichments or exclusions of reference or polymorphic L1 elements are highlighted (light gray). Numbers indicated for the Y chromosome are not reliable, due to its poor sequence coverage (dark gray). (B) Distribution within annotated genes. Simulation again had 3,361,500 “insertion events” distributed randomly genome-wide. The P-value for the comparison of reference L1s inside genes, vs. simulated events inside genes, is <1 × 10−100. The P-value for polymorphic L1s inside genes vs. simulated events inside genes is 2.13 × 10−83 (highlighted gray).