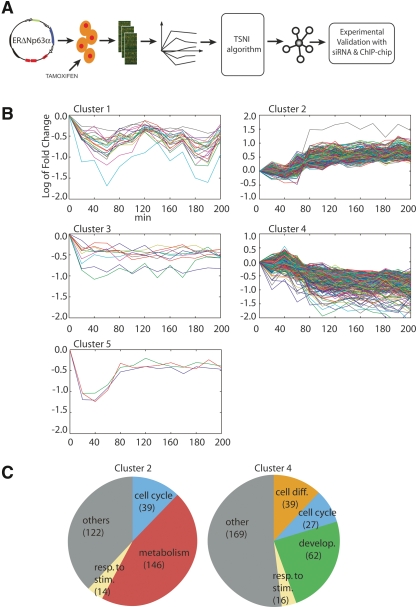
Figure 1.
(A) Experimental and computational approach for the identification of TRP63 target genes. ΔNp63α was fused to a modified estrogen receptor domain (ERΔNp63α) and expressed in primary mouse keratinocytes by retroviral infection. Upon treatment with estrogen agonist tamoxifen, total RNA was collected at 10-min intervals for the first hour, and then at 20-min intervals until 4 h. Dynamic gene expression data were filtered by the TSNI algorithm to yield a ranked list of predicted direct TRP63 target genes. Validation was performed by analyzing global gene expression data upon Trp63 knockdown and by ChIP-chip analysis using TRP63 specific antibodies. (B) Expression profiles of a ΔNp63α responsive gene following its activation. Y-axis shows mRNA levels in the ERΔNp63α expressing keratinocytes treated with tamoxifen versus untreated ones expressed as log2 ratio (i.e., +1 corresponds to a twofold increase with respect to the 0 time point [untreated], −1 to a twofold decrease). Gene expression data were clustered using a Hierarchical Clustering approach with correlation metric and average linkage to generate the hierarchical tree. The number of clusters was set to 5. (C) Transcripts in Clusters 2 and 4 were classified using a functional annotation enrichment analysis (see Supplemental Table 3). The biological categories are as indicated in the pie charts, and the number of transcripts for each category is indicated in parenthesis.