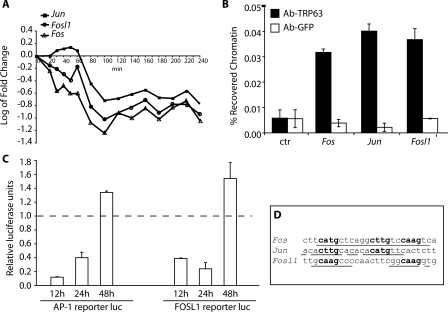
Figure 6.
TRP63 directly regulates AP-1 encoding genes. (A) Expression profiles of AP-1 family members following tamoxifen-induced ERΔNp63α activation in primary keratinocytes. Data for Fosl1 and Jun are the average of two different probes on the microarray (see Supplemental Table 1 for profiles of individual probes). The experiment was performed as described in Figure 2. (B) Specific binding of endogenous TRP63 to Fos, Jun, and Fosl1 promoters. Primary mouse keratinocytes were processed for ChIP with antibodies specific for TRP63 (Ab-TRP63) or unrelated anti-Erk antibodies as control (Ab-GFP), followed by real-time PCR amplification on the promoter sequences indicated in D. Unprecipitated chromatin preparations were similarly analyzed and used as “input” control. The amount of precipitated DNA was calculated relative to the total input chromatin, and expressed as the percentage of the total (Frank et al. 2001). (C) Hela cells were cotransfected with plasmids encoding ΔNp63α, or an empty control vector, together with the luciferase reporter plasmids, Fosl1-Luc, and pAP-1-Luc, carrying multiple copies of the AP-1-Responsive Element. Cells were collected at 12, 24, and 48 h after transfection. Luciferase activity is expressed relative to the empty control vector. Renilla reporter was used for internal normalization. The experiment was repeated twice with similar results. (D) TRP63 binding sites on the Fos, Jun, and Fosl1 promoter regions. The predicted TRP63-binding site is indicated, together with its precise nucleotide sequence: bold nucleotides correspond to the core nucleotide sequence required for TRP63-binding, while underlined nucleotides are matches in the consensus.