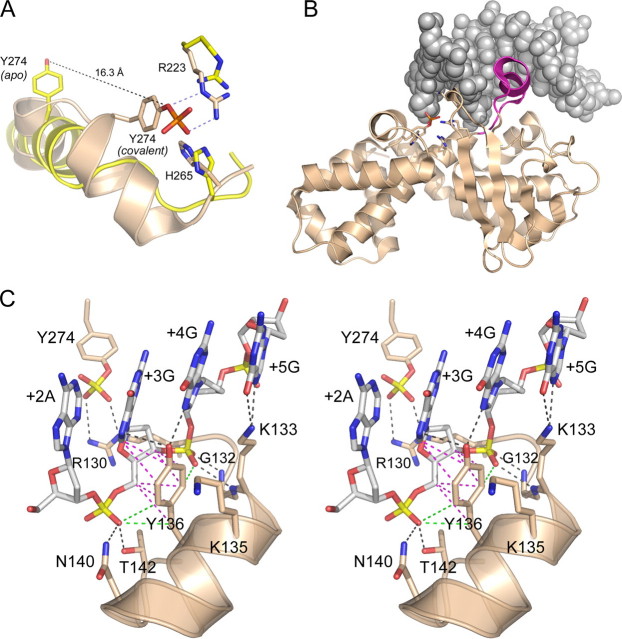
FIGURE 2.
Structural insights to DNA recognition by poxvirus TopIB. A, apoenzyme structure of poxvirus TopIB (PDB 1a41 colored yellow) was superimposed on the covalent tyrosyl-DNA intermediate (PDB 2h7f; colored beige). Catalytic residues Arg223 and His265 overlap in both structures. The image highlights the large movement of Tyr274 and the rearrangement of the flanking secondary structure that is required to bring the nucleophile into the active site for attack on the scissile phosphodiester. B, fold of the C-terminal catalytic domain of poxvirus TopIB in the covalent intermediate is depicted as a ribbon diagram. The duplex DNA flanking the cleavage site is shown as a space-filling model. The image highlights the docking of the specificity helix (colored magenta) in the DNA major groove of the CCCTT recognition site. This segment is disordered in the poxvirus TopIB apoenzyme. C, stereo view of the specificity helix and its interactions with the nonscissile strand 3′-G+5G+4G+3A+2A+1. Ionic and hydrogen bonding interactions are indicated by black dashed lines. Van der Waals contacts of Tyr136 are depicted as magenta dashed lines. Close nonpolar contacts of Tyr136 to the +3 and +4 phosphates are rendered as green dashed lines.