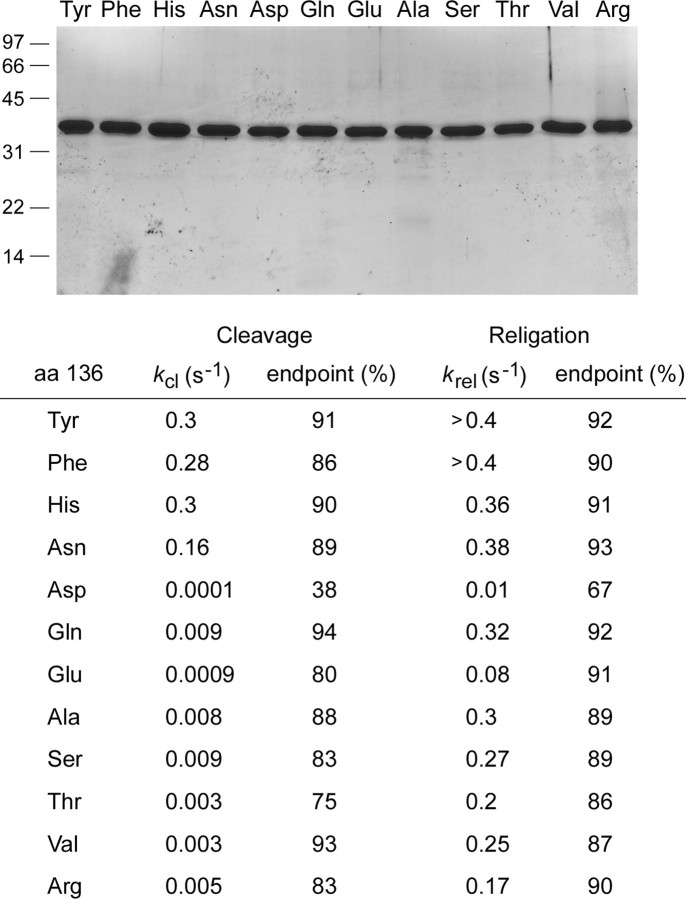
FIGURE 6.
Effects of amino acid substitutions for Tyr136. Top panel, aliquots (4 μg) of the phosphocellulose preparations of vaccinia TopIB with the indicated amino acids at position 136 were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. Bottom panel, single-turnover transesterification. Cleavage reaction mixtures containing (per 20 μl) 50 mm Tris-HCl (pH 7.5), 0.3 pmol of 5′ 32P-labeled 18-mer/30-mer DNA substrate, and 75 ng (2 pmol) of vaccinia TopIB were incubated at 37 °C. The reactions were initiated by adding TopIB to prewarmed reaction mixtures. Aliquots (20 μl) were withdrawn at various times and quenched immediately with SDS (1% final concentration). The samples were digested with proteinase K and then analyzed by urea-PAGE as described in Fig. 4. The apparent cleavage rate constants (kcl) and the cleavage reaction end points (percent of input 32P-DNA covalently attached to TopIB) are shown. Single-turnover religation was assayed as follows. Reaction mixtures containing (per 20 μl) 50 mm Tris-HCl (pH 7.5), 0.3 pmol of 5′ 32P-labeled 18-mer/30-mer DNA substrate, and 75 ng of wild-type or mutant TopIB were incubated at 37 °C for either 5 min (Tyr, His, Phe), 15 min (Asn, Gln), 60 min (Ala, Ser, Val), or 240 min (Thr, Arg, Asp, Glu) to form the suicide intermediate. Religation was initiated by the simultaneous addition of NaCl to 0.5 m and a 5′-OH 18-mer acceptor strand d(ATTCCGATAGTGACTACA) to a concentration of 15 pmol/22 μl (a 50-fold molar excess over the input DNA substrate). Aliquots (22 μl) were withdrawn at the various times thereafter and quenched immediately with SDS. A time 0 sample was withdrawn before the addition of the acceptor strand. The samples were digested with proteinase K and then analyzed by urea-PAGE. The apparent religation rate constants (krel) and the religation reaction end points (percent of the covalent intermediate converted into 30-mer) are shown. aa, amino acids.