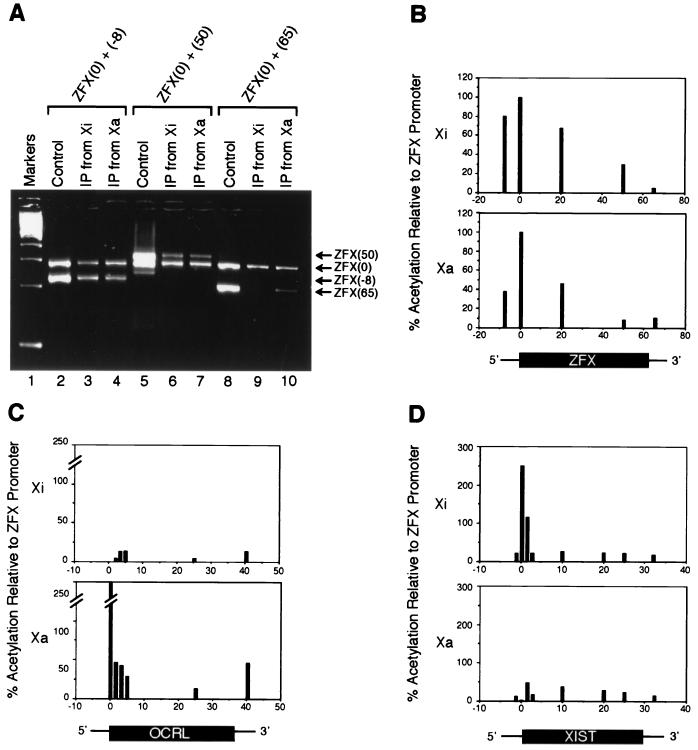
Figure 3.
Quantitative PCR analysis of histone H4 acetylation along X-linked genes. (A) Sample gel of PCR with internal standard used to quantitate relative enrichment of various ZFX sites in chromatin immunoprecipitate. Two sets of primers were used in each PCR: one set, ZFX (0), amplified the ZFX promoter-proximal region and served as an internal standard of comparison, whereas the other set sampled positions along the ZFX genomic locus: ZFX (−8) is 8 kilobases (kb) upstream of the promoter; ZFX (20) is 20 kb downstream of the promoter (data not shown); ZFX (50) is 50 kb downstream of the promoter in the transcribed portion of the gene; and ZFX(65) is 5 kb downstream of the poly(A) site. The control is genomic DNA isolated from cells that were processed in parallel but that were not subjected to IPs. To eliminate differences that were caused by amplification efficiency between primer sets, the amplification ratio from immunoprecipitates was normalized to the ratio obtained with control genomic DNA. After normalization, the relative intensity of the sample PCR product was used to estimate its enrichment relative to ZFX (0) in the immunoprecipitate. (B–D) Graphical representations of relative H4 acetylation levels along X-linked genes as assayed by PCR with an internal standard. Three genes were assayed: ZFX, which escapes X inactivation (B); OCRL, which undergoes X inactivation (C); and XIST, which is transcribed exclusively from Xi (D). The ZFX (0) site was arbitrarily set as the unit of comparison (100%), and acetylation levels at all other sites were expressed as a percentage relative to ZFX (0).