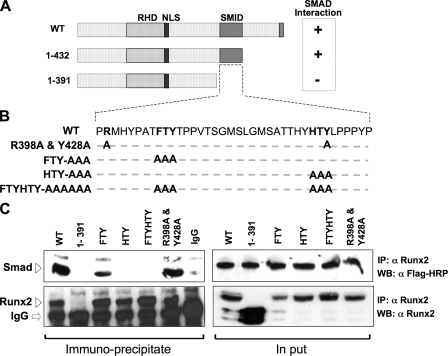
FIGURE 1.
Three residues in the SMID of RUNX2 define physical association with receptor SMADs. A, schematic illustration of the wild-type Runx2 protein with key regulatory domains indicated (RHD, DNA binding runt homology domain; NLS, nuclear localization signal; SMID, Smad interacting domain). SMID was established by deletion mutagenesis and co-immunoprecipitation approach. B, sequence of the 41 amino acids that constitute SMID. Based on SMID, crystal structure residues in two predicted fingers were mutated to alanine. Relative positions of mutated amino acids are indicated in bold. C, HeLa cells were transiently co-transfected with 5 μg of FLAG-tagged receptor Smad1 and HA-tagged wild-type or mutant Runx2 expression plasmid. Cells were treated with 100 ng/ml of recombinant BMP2 3 h post-transfection and harvested 24 h later. Immunoprecipitations were performed on sonicated cell lysates using RUNX2 antibody or species-matched control IgG. Immunoprecipitates and 10% of input samples were resolved on SDS-PAGE, and Western blots were probed with either RUNX2 antibody or FLAG antibody to detect SMAD proteins. Specific bands corresponding to RUNX2 and SMAD 1 are indicated by arrowheads.