Abstract
Mutation in Cu/Zn-superoxide dismutase (SOD1) is a cause of familial amyotrophic lateral sclerosis (ALS). Mutant SOD1 protein (SOD1mut) induces motor neuron death, although the molecular mechanism of SOD1mut-induced cell death remains controversial. Here we show that SOD1mut specifically interacted with Derlin-1, a component of endoplasmic reticulum (ER)-associated degradation (ERAD) machinery and triggered ER stress through dysfunction of ERAD. SOD1mut-induced ER stress activated the apoptosis signal-regulating kinase 1 (ASK1)-dependent cell death pathway. Perturbation of binding between SOD1mut and Derlin-1 by Derlin-1-derived oligopeptide suppressed SOD1mut-induced ER stress, ASK1 activation, and motor neuron death. Moreover, deletion of ASK1 mitigated the motor neuron loss and extended the life span of SOD1mut transgenic mice. These findings demonstrate that ER stress-induced ASK1 activation, which is triggered by the specific interaction of Derlin-1 with SOD1mut, is crucial for disease progression of familial ALS.
Keywords: Amyotrophic lateral sclerosis, endoplasmic reticulum-associated degradation, endoplasmic reticulum stress, Derlin-1, ASK1
Amyotrophic lateral sclerosis (ALS) is the most frequent adult-onset motor neuron disease and is characterized by selective loss of motor neurons. Familial ALS-linked mutations of Cu/Zn-superoxide dismutase (SOD1) induce motor neuron death. Previous studies have suggested that SOD1mut causes various cellular events, including alteration of gene expression (Yoshihara et al. 2002; Kirby et al. 2005), abnormal protein interactions (Kunst et al. 1997), activation of caspases (Pasinelli et al. 1998; Li et al. 2000), dysfunction of mitochondria (Bowling et al. 1993; Wong et al. 1995; Liu et al. 2004), and cytoskeletal abnormalities (Julien and Beaulieu 2000). However, the causal relationship between these events and motor neuron death remains unclear.
Endoplasmic reticulum (ER) stress is triggered by the accumulation of misfolded proteins within the ER lumen and has recently been implicated in various neurodegenerative diseases (Sekine et al. 2006). Recent studies have suggested that ER stress signaling is also involved in the pathogenesis of ALS (Tobisawa et al. 2003; Atkin et al. 2006; Kikuchi et al. 2006). We showed previously that, upon ER stress, an ER-resident type I transmembrane serine/threonine kinase termed IRE1 recruits TRAF2 and ASK1 (apoptosis signal-regulating kinase 1) on the ER membrane and thus activates the ASK1-dependent apoptosis pathway (Nishitoh et al. 2002). Several groups have reported that activation of the ASK1 cascade is associated with induction of motor neuron death by SOD1mut both in vitro (Raoul et al. 2002) and in vivo (Wengenack et al. 2004; Holasek et al. 2005; Veglianese et al. 2006). These observations suggested that a functional link between ER stress and ASK1 may exist in the process of SOD1mut-induced motor neuron death. However, the molecular mechanism by which SOD1mut induces ER stress has remained unclear. Furthermore, genetic evidence has not been provided regarding the hypothetical involvement of ER stress–ASK1 pathway in SOD1mut-induced motor neuron death. In the present study, we investigated the molecular mechanism of SOD1mut-induced ER stress and the role of ER stress-induced ASK1 activation in the pathogenesis of ALS.
Results
SOD1mut triggers ER stress
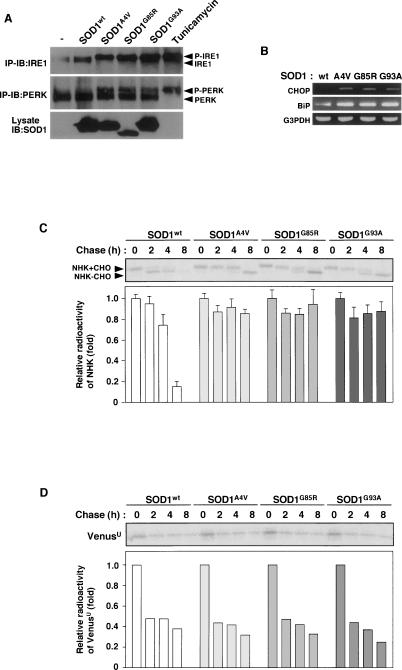
To investigate the causal relationship between SOD1mut and ER stress-dependent motor neuron death, we first examined whether SOD1mut induces ER stress in NSC34 motor neurons, as assessed by band-shift analyses of the ER transmembrane kinase receptors IRE1 and PERK. Adenovirus (Ad)-mediated expression of ALS-linked SOD1mut (SOD1G93A) was detectable within 48 h of infection (Supplemental Fig. S1A). SOD1mut (SOD1A4V, SOD1G85R, and SOD1G93A) but not wild-type SOD1 (SOD1wt) activated IRE1 and PERK (Fig. 1A; Supplemental Fig. S1A). To confirm the activation of IRE1 by SOD1mut, we examined Xbp-1 mRNA splicing by RT–PCR. We clearly observed the appearance of spliced Xbp-1 mRNA by SOD1mut but not SDO1wt in HEK293 cells (Supplemental Fig. S1B) and NSC34 cells (Fig. 4D, lanes 2–5, below). SOD1mut-specific induction of CHOP and BiP (ER stress marker proteins) was also observed (Fig. 1B). These findings suggested that accumulation of various SOD1mut proteins commonly induce ER stress.
Figure 1.
SOD1mut triggers ER stress and inhibition of ERAD. (A) NSC34 cells were lysed after infection with Ad-SOD1wt, Ad-SOD1A4V, Ad-SOD1G85R, or Ad-SOD1G93A for 48 h or treatment with 2.5 μg/mL tunicamycin (a potent inducer of ER stress) for 2 h and analyzed by immunoprecipitation-immunoblotting (IP-IB) with antibodies to IRE1α or PERK. (P-IRE1) Activated IRE1; (P-PERK) activated PERK. The presence of SOD1 in the same lysates is shown. (B) NSC34 cells were transfected with SOD1wt or SOD1mut for 48 h. Expression of BiP and CHOP was examined by RT–PCR. (C) NSC34 cells were transfected with NHK and SOD1wt or SOD1mut. Cells were pulse-labeled with [35S] methionine and cysteine and chased for the indicated time periods. Cell lysates were immunoprecipitated with antibody to α1AT. (NHK + CHO) Glycosylated NHK; (NHK − CHO) deglycosylated NHK. The relative radioactivities in NHK at different times of chase were calculated and are shown as fold decreases relative to the intensity observed at 0 h chase. (D) NSC34 cells were transfected with VenusU and SOD1wt or SOD1mut. Cells were pulse-labeled with [35S] methionine and cysteine and chased for the indicated time periods. Cell lysates were immunoprecipitated with antibody to GFP. The relative radioactivities in VenusU at different times of chase were calculated and are shown as fold decreases relative to the intensity observed at 0 h chase.
Figure 4.
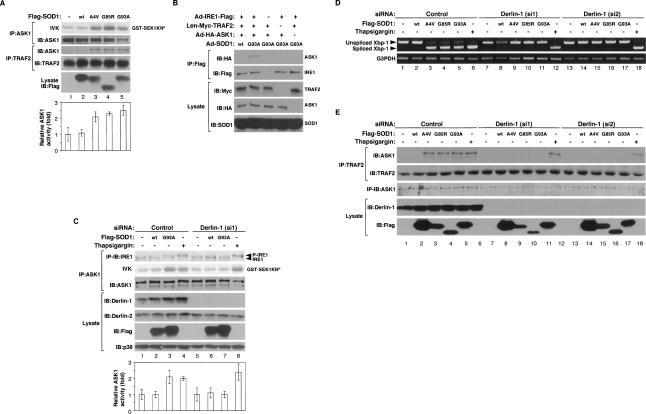
SOD1mut activates the IRE1–TRAF2–ASK1 pathway. (A) NSC34 cells were infected with Ad-SOD1wt, Ad-SOD1A4V, Ad-SOD1G85R, or Ad-SOD1G93A for 48 h. ASK1 activity was measured by in vitro kinase assay (IVK). (GST-SEK1KNP) Phosphorylated GST-SEK1KN. Interaction between TRAF2 and ASK1 was analyzed by IP with antibody to TRAF2 and IB with antibody to ASK1. The presence of ASK1, TRAF2, and SOD1 in the same lysates is shown. (Lane 1) Kinase activity relative to the amount of ASK1 protein is shown as fold increase compared with control cells. (B) NSC34 cells were infected with adenoviruses and lentivirus at the indicated combinations. SOD1mut-dependent IRE1–TRAF2–ASK1 complex formation were analyzed by IP-IB. (C) NSC34 cells, transfected with siRNA against Derlin-1 (si1) or nonspecific sequence (Control), were infected with Ad-SOD1wt or Ad-SOD1G93A for 48 h or treatment with 10 μM Thapsigargin for 2 h. Activation of IRE1 and ASK1 was analyzed by IP-IB with antibodies to IRE1α and by IVK using GST-SEK1KN as a substrate, respectively. The presence of ASK1, Derlin-1, Derlin-2, SOD1, and p38 in the same lysates is shown. (Lanes 1,5) Kinase activity relative to the amount of ASK1 protein is shown as fold increase compared with control cells. (D,E) NSC34 cells, transfected with siRNA against Derlin-1 (si1), Derlin-1 (si2), or nonspecific sequence (Control), were infected with Ad-SOD1wt, Ad-SOD1A4V, Ad-SOD1G85R, or Ad-SOD1G93A for 48 h or treatment with 2 μM thapsigargin for 2 h. Xbp-1 mRNA splicing was determined by RT–PCR. Interaction between TRAF2 and ASK1 was analyzed by IP with antibody to TRAF2 and IB with antibody to ASK1. The presence of TRAF2, ASK1, Derlin-1, and Flag-SOD1 in the same lysates is shown.
Inhibition of proteasome activity has been reported to be induced by SOD1mut in Neuro2a cells (Urushitani et al. 2002). Since polyglutamine (polyQ) fragments have been shown to induce ER stress through inhibition of proteasome activity (Nishitoh et al. 2002), SOD1mut might also induce ER stress through proteasomal dysfunction. We therefore examined whether alteration of proteasome activity was involved in SOD1mut-induced ER stress. Proteasome activity was inhibited by treatment with lactacystin, a proteasome inhibitor, but not by Ad-mediated expression of SOD1mut in NSC34 cells within 48 h (Supplemental Fig. S2A). These findings suggested that a mechanism other than proteasomal dysfunction may be involved in SOD1mut-induced ER stress in NSC34 cells.
SOD1mut inhibits ERAD
Restoration of ER homeostasis is achieved mainly by two independent mechanisms, chaperone-dependent refolding and ER-associated degradation (ERAD). In ERAD, misfolded proteins are exported from the ER back into the cytosol and are rapidly degraded by the ubiquitin–proteasome system (UPS) (Kopito 1997; Tsai et al. 2002; Meusser et al. 2005). Blocking of the refolding mechanism and/or the ERAD mechanism induces accumulation of misfolded proteins within the ER lumen and thus ER stress. Since SOD1mut clearly induced expression of the ER-resident chaperone BiP (Fig. 1B), the productive refolding mechanism appears not to be inhibited by SOD1mut. To examine whether SOD1mut interferes with the ERAD mechanism, we examined the stability of the null Hong Kong (NHK) mutant protein of α1-antitrypsin (α1AT), an ER luminal misfolded protein (Sifers et al. 1988). Pulse-chase experiments showed that NHK was degraded with a half-life of <8 h in NSC34 cells transfected with or without SOD1wt (Fig. 1C; Supplemental Fig. S3A). In contrast, overexpression of SOD1mut decreased the degradation of NHK to a half-life of >8 h (Fig. 1C). To confirm the interference with the degradation of the other ERAD substrate by SOD1mut, we examined the stability of CD3δ, a well-characterized transmembrane-type ERAD substrate (Fang et al. 2001). Cycloheximide chase experiments showed that the degradation of CD3δ was also retarded by overexpression of SOD1mut (Supplemental Fig. S3C). To further examine whether SOD1mut also interferes with the degradation of cytosolic protein, we examined the stability of the unstable Venus mutant protein (VenusU), a cytosolic proteasome substrate. There was no difference between a half-life of VenusU in SOD1wt transfected cells and that in SOD1mut transfected cells (Fig. 1D). These findings suggested that SOD1mut specifically impairs ERAD function but not cytosolic protein degradation in motor neurons. Furthermore, SOD1mut clearly delayed the deglycosylation of NHK in addition to degradation (see the band shift of NHK; Fig. 1C). Previous studies have reported that the cytoplasmic peptide N-glycanase contributes to deglycosylation of misfolded glycoproteins prior to degradation (Suzuki et al. 2002). Thus, the delay of the deglycosylation of NHK suggested that SOD1mut delays retro-translocation from ER lumen to the cytosol.
SOD1mut interacts specifically with Derlin-1
Since inhibition of the UPS is unlikely to be essential for SOD1mut-induced ER stress (Fig. 1D; Supplemental Fig. S2A), disturbance of the retro-translocation system from the ER lumen to the cytoplasm may be a potential target of SOD1mut. Various proteins involved in ERAD have been identified recently (Meusser et al. 2005). Among them, components of the retro-translocation machinery including ATPase p97, its cofactors Ufd1 and Npl4, and the ER membrane proteins Derlin-1 and VIMP are of key importance to ERAD function (Lilley and Ploegh 2004; Ye et al. 2004). We therefore examined the in vitro interactions of SOD1mut with p97, Ufd1, Npl4, VIMP, and Derlin-1. In vitro-translated 35S-labeled Derlin-1, but not p97, Ufd1, Npl4, or VIMP, bound specifically to recombinant His-tagged SOD1G93A (Fig. 2A). To examine the interaction of Derlin-1 with different mutant forms of SOD1, several SOD1 mutants, including the A4V, G85R, and G93A mutants, were analyzed in mammalian cells. SOD1A4V, SOD1G85R, and SOD1G93A, but not SOD1wt, were clearly coimmunoprecipitated with Derlin-1 (Fig. 2B). The remaining ERAD components, including p97, Npl4, Ufd1, and VIMP, were not coimmunoprecipitated with SOD1mut (data not shown). The specific interaction between exogenous SOD1mut and endogenous Derlin-1 was also observed in NSC34 cells (Fig. 2C). These findings suggested that SOD1mut proteins specifically interact with Derlin-1 in mammalian cells.
Figure 2.
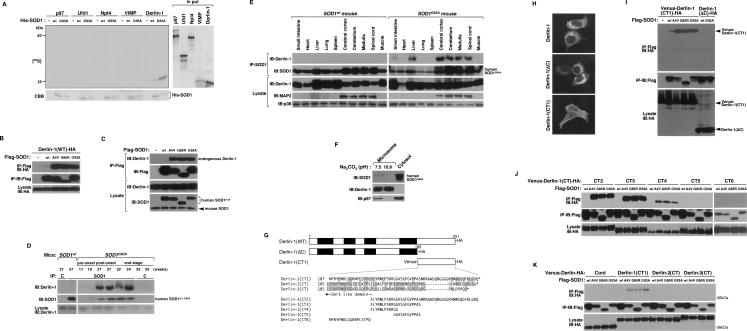
SOD1mut interacts with Derlin-1. (A) 35S-labeled p97, Ufd1, Npl4, VIMP, or Derlin-1 proteins were incubated with recombinant His-SOD1wt or -SOD1G93A proteins and immobilized on Ni-NTA beads (top left). The bottom part of the SDS-PAGE gel was stained with Coomassie brilliant blue dye (CBB). Amounts of incubated 35S-labeled proteins are shown (input). (B) Lysates from HEK293 cells, transfected at the indicated combinations, were analyzed by IP-IB. (C) NSC34 cells were lysed after infection with Ad-Flag-SOD1wt, Ad-Flag-SOD1A4V, Ad-Flag-SOD1G85R, or Ad-Flag-SOD1G93A for 48 h and analyzed by IP-IB. (D) Extracts from spinal cords of mice were immunoprecipitated with an antibody to SOD1 (IP: SOD1) or control nonimmune antibody (IP: C) and analyzed by IB with antibodies to Derlin-1 and SOD1. The presence of Derlin-1 in the same lysates is shown. (E) Extracts from tissues of SOD1wt mice and SOD1G93A mice were analyzed by IB with antibodies to Derlin-1, MAP2, or p38. Interaction between SOD1mut and Derlin-1 in tissues of SOD1G93A mice was analyzed by IP-IB. (F) Spinal cord of SOD1G93A mouse was homogenized and the microsomes were isolated from the post-nuclear and post-mitochondria homogenate. For alkaline extraction, the microsomes were incubated in 100 mM Na2CO3 (pH 7.5 or pH 10.9). After the incubation, the microsomes were pelleted and analyzed by IB with antibodies to Derlin-1, SOD1, and p97. Derlin-1 and p97 were used as controls for ER membrane-anchored protein and peripheral protein, respectively. (G) Schematic representation of various mutant forms of Derlin-1. Transmembrane domains of Derlin-1 are shown by black boxes. Identical residues of C-terminal fragment of Derlin family proteins are shaded. (H) HEK293 cells were transfected with various mutant forms of Derlin-1 and stained with antibody to HA. (I–K) Lysates from HEK293 cells, transfected at the indicated combinations, were analyzed by IP-IB.
To confirm these results in more physiological conditions, we examined the association of endogenous Derlin-1 and SOD1 in spinal cord of SOD1G93A gene transgenic (SOD1G93A) mice. SOD1G93A mice showed the onset of motor function loss, which was measured using rota-rod test, at a mean age of 28.1 ± 1.3 wk, and died at a mean age of 34.9 ± 1.6 wk (see below and Fig. 5G). Accumulation of SOD1mut was observed in the spinal cord of SOD1G93A mice at the post-onset stage and the end stage (Fig. 2D). A strong interaction of endogenous Derlin-1 with exogenous SOD1G93A, but not with exogenous SOD1wt (Supplemental Fig. S8A), was observed in parallel with accumulation of SOD1mut (Fig. 2D).
Figure 5.
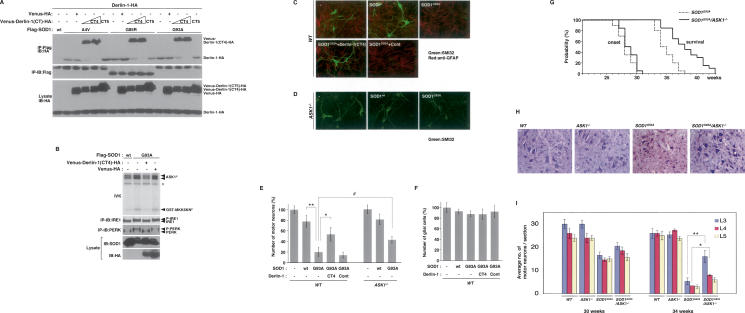
Impairment of SOD1mut–Derlin-1 interaction and deletion of ASK1 mitigate SOD1mut-induced motor neurotoxicity. (A) Lysates from HEK293 cells, transfected at the indicated combinations, were analyzed by IP-IB. (B) NSC34 cells were infected with Ad-SOD1wt, Ad-SOD1G93A, Len-Venus-Derlin-1(CT4)-HA, and Len-Venus-HA at the indicated combinations for 48 h. Activation of IRE1 and PERK was examined as described in Figure 1A. Activation of ASK1 was analyzed by IVK using GST-MKK6KN as substrate. (ASK1P) Autophosphorylated ASK1. (GST-MKK6KNP) Phosphorylated GST-MKK6KN. Asterisk denotes nonspecific bands. (C) Wild-type mice spinal cord cultures were infected with lentivirus at the indicated combinations for 72 h. Cultures were fixed and doubly stained with antibodies to nonphosphorylated neurofilament (SMI32) (green) and GFAP (red). (D) ASK1−/− mice spinal cord cultures were infected as indicated and stained with SMI32 antibody. (E) The percentage of total cell count of SMI32 antibody-positive cells is shown compared with control culture (wild type; n = 3); (ASK1−/−; n = 5). Values are means ± SE of independent experiments. (*) P < 0.05; (**) P < 0.01; significance calculated by Student’s t-test. (#) P < 0.05 significance calculated by ANOVA. (F) The percentage of total cell count of anti-GFAP antibody-positive cells derived from wild-type mice is shown compared with control culture. Values are means ± SE of three independent experiments. (G) The onset of disease was determined by motor function deficit seen in rota-rod performance in SOD1G93A mice in the presence or absence (ASK1−/−) of ASK1. The cumulative probability of onset of rota-rod deficit was not significantly changed in SOD1G93A/ASK1−/− mice (n = 10; solid line) compared with SOD1G93A mice (n = 10; dotted line). Probabilities of survival reveal prolongation of life span of SOD1G93A/ASK1−/− mice (n = 20; solid line) compared with SOD1G93A mice (n = 20; dotted line). The data were analyzed by the Kaplan-Meier life test and by long-rank test. (H) Cresyl violet (Nissl)-stained paraffin sections of ventral horn from lumbar (level L3) spinal cords at end stage (age 34 wk) are shown. (I) Stereological analysis of motor neuron death. Numbers of motor neurons were determined by counting the large Nissl-positive neurons in the ventral horn. Five mice of each group were used for analysis. Thirty-week-old and 34-wk-old mice were sacrificed, and sections of lumbar spinal cord at levels L3, L4, and L5 were counted for each mouse. Values are means ± SE. (*) P < 0.05; (**) P < 0.01; significance calculated by Student’s t-test.
The reported expression pattern of Derlin-1 in mouse organs is controversial (Lilley and Ploegh 2005; Oda et al. 2006). We thus examined whether Derlin-1 is expressed in neuronal tissues of SOD1wt and SOD1G93A mice by immunoblotting analysis using a newly developed Derlin-1-specific antibody (Supplemental Fig. S4A). Derlin-1 proteins were clearly observed in many organs, including cerebral cortex, cerebellum, medulla, and spinal cord (Fig. 2E). Since human SOD1G93A gene is driven by human SOD1 promoter, SOD1G93A was widely expressed as expected in SOD1G93A mouse tissues (Fig. 2E; Epstein et al. 1987; Gurney et al. 1994). We also found that expression of SOD1G93A in neuronal tissues and liver was higher than that in other organs and that the interaction of Derlin-1 with SOD1mut, but not SOD1wt, was observed specifically in these tissues (Fig. 2E). The regionally specific interaction of Derlin-1 with SOD1mut might thus be one of the reasons why SOD1mut specifically targets neuronal tissues.
SOD1mut interacts with the C-terminal cytoplasmic region of Derlin-1
Although SOD1mut itself has neither a signal sequence nor transmembrane segment and is located mainly in the cytosol, it has been reported that SOD1mut accumulates in the ER (Kikuchi et al. 2006; Urushitani et al. 2006). We therefore examined whether SOD1mut resides in the ER lumen or associates with the cytosolic side of the ER membrane. Purified microsomal fraction enriched in the ER membrane was isolated from the spinal cord of a SOD1G93A mouse. We found that SOD1G93A exists mainly in the cytosolic fraction and some in the microsomal fraction (Fig. 2F). The alkaline extraction clearly released SOD1G93A, but not Derlin-1, from the microsome, as seen for p97, which is known to be peripherally associated with the ER membrane (Fig. 2F). These findings indicated that SOD1mut exists mainly in the cytosol, and some fraction of SOD1mut attaches to the cytosolic surface of the ER.
Derlin-1 contains a domain named for yeast Der1p (Der1-like domain) and is a protein that spans the ER membrane four times, with both its N and C termini facing the cytosol (Fig. 2G; Lilley and Ploegh 2004; Ye et al. 2004). To determine the domain of interaction, we constructed expression plasmids for a C-terminally truncated Derlin-1 protein [Derlin-1(ΔC)], which exhibited a reticular pattern, typical of ER localization, and for a C-terminal fragment of Derlin-1 [Derlin-1(CT1)], which exhibited cytosolic localization (Fig. 2H). Derlin-1(CT1), but not Derlin-1(ΔC), coimmunoprecipitated with SOD1mut (Fig. 2I), indicating that SOD1mut associates with the C-terminal cytosolic region of Derlin-1. To further investigate the region of interaction of Derlin-1(CT1) with SOD1mut, various truncation mutants of the C-terminal fragment of Derlin-1 were assessed for interaction with SOD1mut. Derlin-1(CT1), Derlin-1(CT2), Derlin-1(CT3), and Derlin-1(CT4), but not Derlin-1(CT5) and Derlin-1(CT6), were coimmunoprecipitated with SOD1mut (Fig. 2J), indicating that Derlin-1(CT4), composed of 12 amino acids (FLYRWLPSRRGG), is minimally required and sufficient for interaction with SOD1mut. Mammals also express two additional Der1-like homologs to Derlin-1, designated Derlin-2 and Derlin-3 (Oda et al. 2006). To examine whether SOD1mut also interacts with other Derlin family proteins, C-terminal fragments of Derlin-2 and Derlin-3 [Derlin-2(CT) and Derlin-3(CT), respectively] were assessed for interaction with SOD1mut. Derlin-2(CT) and Derlin-3(CT) were not coimmunoprecipitated with SOD1mut (Fig. 2K), suggesting that, among the Derlin family proteins, SOD1mut associates specifically with Derlin-1. Moreover, since mRNA expression of Derlin-1 was more abundant than that of Derlin-2 and Derlin-3 in brain (Oda et al. 2006), SOD1mut in neuronal tissues probably targets ERAD through specific binding to Derlin-1.
SOD1mut attenuates the retro-translocation of ERAD substrates on the components of ERAD machinery
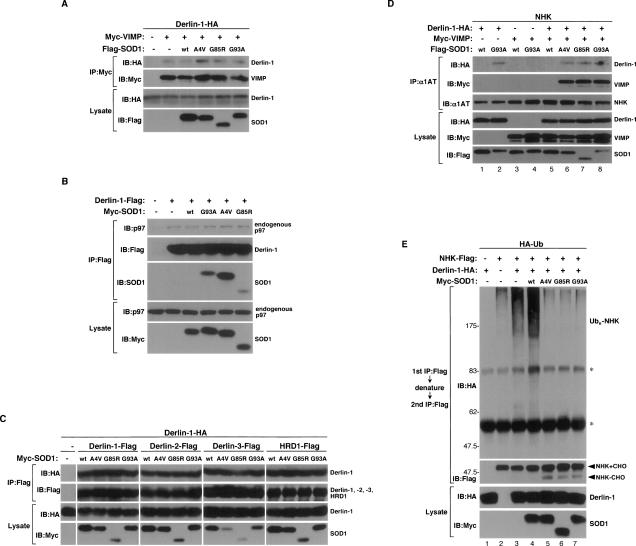
There are at least three models for the mechanism of how ERAD is inhibited by SOD1mut: (1) Since the C-terminal domain of Derlin-1 has been shown to be necessary for the recruitment of p97 on the ER membrane through binding with VIMP (Ye et al. 2004; Lilley and Ploegh 2005), SOD1mut might inhibit the assembly of Derlin-1, VIMP, and p97 on the ER membrane; (2) since Derlin-1 has been reported to form homo- as well as hetero-oligomers and the ERAD complex with HRD1, an ER-anchored ubiquitin E3 ligase (Lilley and Ploegh 2005; Schulze et al. 2005; Ye et al. 2005; Oda et al. 2006), SOD1mut might inhibit the interaction of Derlin-1 with these ERAD components; (3) SOD1mut might inhibit the directional flow of ERAD substrates at a certain step of their transfer from the ER lumen to p97 or E3 ligase. If the first two models were correct, overexpression of SOD1mut would inhibit the coimmunoprecipitation of Derlin-1 with VIMP, p97, or the other ERAD components. However, we could not detect any dissociation of VIMP (Fig. 3A), p97 (Fig. 3B), Derlin family proteins, and HRD1 (Fig. 3C) from Derlin-1 by SOD1mut. To test model 3, we examined the interaction of an ERAD substrate (NHK) with Derlin-1. SOD1mut, but not SOD1wt, induced the interaction between NHK and Derlin-1 (Fig. 3D, top panel, lanes 2,6–8). Furthermore, NHK was found to associate with VIMP in the presence of Derlin-1 and SOD1mut (Fig. 3D, second panel, lanes 6–8), suggesting that SOD1mut induces formation of an NHK–Derlin-1–VIMP complex on the ER membrane.
Figure 3.
SOD1mut attenuates the retro-translocation of ERAD substrate on the components of ERAD. (A) Lysates from HEK293 cells, transfected with Derlin-1-HA, Myc-VIMP, and Flag-SOD1 at the indicated combinations, were analyzed by IP-IB. The presence of Derlin-1-HA and Flag-SOD1 in the same lysates is shown. (B) Lysates from HEK293 cells, transfected at the indicated combinations, were analyzed by IP-IB. The presence of p97 and Myc-SOD1 in the same lysates is shown. (C) Lysates from HEK293 cells, transfected at the indicated combinations, were analyzed by IP-IB. The presence of Derlin-1-HA and Myc-SOD1 in the same lysates is shown. (D) Lysates from HEK293 cells, transfected with NHK, Derlin-1-HA, Myc-VIMP, and Flag-SOD1 at the indicated combinations, were analyzed by IP-IB. The presence of Derlin-1-HA, Myc-VIMP, and Flag-SOD1 in the same lysates is shown. (E) HEK293 cells were transfected with NHK-Flag, Derlin-1-HA, Myc-SOD1, and HA-Ub at the indicated combinations and incubated with 0.25 μM MG132 for 18 h. NHK was immunoprecipitated with antibody to Flag. After incubation with the denaturing buffer containing 1% SDS, NHK was reimmunoprecipitated with antibody to Flag. Samples were immunoblotted with antibodies to HA and Flag. The presence of Derlin-1-HA and Myc-SOD1 in the same lysates is shown. Asterisks denote nonspecific bands and IgG.
The ERAD substrate that emerged into the cytosol is captured by p97, polyubiquitinated by E3 ligase, and degraded by the proteasome (Tsai et al. 2002; Meusser et al. 2005). We next examined whether SOD1mut inhibits ubiquitination of NHK. Overexpression of Derlin-1 increased the amount of ubiquitinated NHK in the presence of proteasome inhibitor (MG132) (Fig. 3E, top panel, lane 3), suggesting that Derlin-1 contributes to the degradation of NHK. Expression of SOD1mut clearly decreased the amount of ubiquitinated NHK (Fig. 3E, top panel, lanes 5–7). Furthermore, SOD1mut induced accumulation of nonubiquitinated and deglycosylated NHK in addition to glycosylated NHK (Fig. 3E, second panel, lanes 5–7), suggesting that SOD1mut blocks the presentation of NHK from retro-translocon to E3 ligase. These data also suggested that SOD1mut does not inhibit the assembly of essential components of retro-translocon per se but does trap ERAD substrates on the complex composed of SOD1mut–Derlin-1–VIMP and thereby inhibits subsequent transfer of ERAD substrates to the ubiquitination step by E3.
SOD1mut activates the IRE1–TRAF2–ASK1 pathway
It has been reported that the ASK1–p38 pathway is activated in motor neurons of the spinal cord of SOD1G93A mice (Wengenack et al. 2004; Holasek et al. 2005; Veglianese et al. 2006). To investigate the role of ASK1 in the motor neurotoxicity by SOD1mut, we examined whether SOD1mut activates ASK1 as assessed by in vitro kinase assay. Expression of SOD1mut, but not SOD1wt, activated endogenous ASK1 (Fig. 4A, top panel). We next examined whether SOD1mut-induced ASK1 activation is mediated by ER stress. Activated IRE1 has been demonstrated to recruit TRAF2 and ASK1 on the ER membrane and thus to activate ASK1 (Nishitoh et al. 2002). We examined whether SOD1mut induces interaction between endogenous ASK1 and TRAF2. ASK1 was found to associate with TRAF2 in NSC34 cells infected with Ad-SOD1mut but not those infected with Ad-SOD1wt (Fig. 4A, third panel). To further examine whether IRE1 recruits TRAF2 and ASK1 in SOD1mut-expressing cells, Ad-SOD1, Ad-ASK1, Ad-IRE1, and Lentivirus encoding (Len)-TRAF2 were infected into NSC34 cells and subjected to coimmunoprecipitation analysis. ASK1 was found to associate with IRE1 only in the presence of TRAF2 and SOD1mut (Fig. 4B), suggesting that SOD1mut induces formation of an IRE1–TRAF2–ASK1 complex on the ER membrane and thus activates ASK1 by triggering ER stress-induced IRE1 activation.
We next assessed the requirement of Derlin-1 for SOD1mut-induced ER stress and ASK1 activation using siRNA against Derlin-1. Derlin-1 siRNA effectively suppressed expression of Derlin-1 without affecting that of Derlin-2, ASK1, p38, or TRAF2 (Fig. 4C,E). We examined whether reducing Derlin-1 expression levels affected the ER stress response using band-shift analyses of IRE1 and Xbp-1 mRNA splicing, which is caused by the activated RNase domain of IRE1. Derlin-1 siRNA exhibited no effect on the basal level of activation of IRE1 (Fig. 4C [lanes 1,5], D [lanes 1,7,13]) and thapsigargin-induced activation of IRE1 (Fig. 4C [lanes 4,8], D [lanes 6,12,18]), suggesting that Derlin-1 is not essential for ERAD in NSC34 cells. Interestingly, however, SOD1mut-induced activation of IRE1 and ASK1 was clearly inhibited by Derlin-1 depletion (Fig. 4C [lane 7], D [lanes 9–11,15–17]). Furthermore, SOD1mut-induced interaction between TRAF2 and ASK1 was inhibited by Derlin-1 depletion (Fig. 4E, lanes 9–11,15–17), suggesting that Derlin-1 is selectively required for the SOD1mut-induced ER stress. These results also suggested that SOD1mut induces ERAD dysfunction not by a simple loss of function but by a gain of malfunction of Derlin-1.
Dissociation of SOD1mut from Derlin-1 protects motor neurons from SOD1mut-induced cell death
Based on the above findings, which indicate that SOD1mut proteins induce ER stress by interacting with Derlin-1, we hypothesized that forced dissociation of SOD1mut from Derlin-1 may restore normal function of ERAD and thus suppress SOD1mut-induced motor neurotoxicity. We examined the effect of Derlin-1(CT4) on the interaction between SOD1mut and Derlin-1. When Derlin-1(CT4) but not control (Venus-HA) or Derlin-1(CT5) was coexpressed, binding of Derlin-1 to SOD1mut was strongly inhibited (Fig. 5A). Reciprocally, Derlin-1(CT4) was found to associate with SOD1mut in a dose-dependent fashion (Fig. 5A). We next examined the effect of Derlin-1(CT4) on SOD1mut-induced ER stress and ASK1 activation. Derlin-1(CT4) inhibited SOD1mut-induced activation of IRE1, PERK, and ASK1 (Fig. 5B). These findings suggested that the interaction between SOD1mut and Derlin-1 plays a central role in the activation of the ER stress–ASK1 pathway.
We next investigated the effects of Derlin-1(CT4) on SOD1mut-induced cell death using spinal cord cultures derived from E12.5 mouse embryos (Urushitani et al. 2006). At 7 d after plating, spinal cord cultures were infected with Len-SOD1wt or Len-SOD1mut with Len-Derlin-1(CT4) or control (Venus-HA) lentivirus for additional 3 d. Expression of neither SOD1wt nor SOD1G93A affected the number of glial cells (Fig. 5C,F). In contrast, the number of motor neurons that were stained with antibody to unphosphorylated neurofilament-H (SMI32) was strongly decreased by SOD1G93A (Fig. 5C,E) and SOD1G85R (Supplemental Fig. S5A,B), indicating that SOD1mut was selectively toxic to motor neurons and not glial cells. SOD1mut-induced motor neuron death was significantly attenuated by coexpression of Derlin-1(CT4) (Fig. 5C,E; Supplemental Fig. S5A,B). These findings are consistent with the inhibitory effects of Derlin-1(CT4) on the binding between SOD1mut and Derlin-1 and on the ER stress-mediated activation of ASK1, and thus strongly suggested that induction of dysfunction of Derlin-1 by SOD1mut may contribute to the pathogenesis of familial ALS.
ASK1 deficiency mitigates motor neuron death and prolongs survival of ALS mice
Finally, we examined the requirement of ASK1 for SOD1mut-induced motor neuron death. Spinal cord cultures derived from wild-type and ASK1−/− mice were infected with Len-SOD1wt or Len-SOD1G93A. ASK1 deficiency did not affect the expression of SOD1wt or SOD1G93A (Supplemental Fig. S6). ASK1−/− neurons were significantly more resistant to SOD1mut-induced cell death than were wild-type neurons (Fig. 5D,E). Inhibition of SOD1mut-induced cell death in ASK1−/− neurons was not significantly enhanced by the expression of Derlin-1(CT4) (Supplemental Fig. S7), suggesting that the ASK1-dependent signal may serve as a major cell death pathway in the downstream SOD1mut-induced ER stress. To further evaluate the role of ASK1 in the pathogenesis of ALS in vivo, we tested whether ASK1 deficiency ameliorates the motor neuropathological alterations in ALS mice. We produced SOD1G93A mice that lacked ASK1 (SOD1G93A/ASK1−/− mice) (Supplemental Fig. S8B) and compared their time of disease onset and life span with those of SOD1G93A mice. Time of disease onset was determined as that of loss of motor function. On rota-rod testing, no significant difference was found between SOD1G93A and SOD1G93A/ASK1−/− mice (Fig. 5G). However, the mean survival of SOD1G93A/ASK1−/− mice was 38.4 ± 2.7 wk (±SEM) and significantly longer than the 34.9 ± 1.6 wk survival of control SOD1G93A mice (long rank = 17.0206, P < 0.0001) (Fig. 5G). We also examined the time of disease onset and life span using SOD1G93A high copy [SOD1G93A(high)] mice. ASK1 deficiency also extended the survival of SOD1G93A(high) mice but not the time of onset (Supplemental Fig. S9). These findings indicated that ASK1 deficiency extends the survival of ALS mice by ameliorating disease progression. To assess the role of ASK1 in spinal motor neuron death in vivo, we examined L3, L4, and L5 spinal sections from wild-type, ASK1−/−, SOD1G93A, and SOD1G93A/ASK1−/− mice. At the end stage, SOD1G93A mice exhibited fewer motor neurons in the anterior horn of the spinal cord than age-matched wild-type or ASK1−/− mice (Fig. 5H; Supplemental Fig. S10A). In contrast, a larger number of motor neurons were significantly found in the upper (L3 and L4) spinal cord from SOD1G93A/ASK1−/− than in that from SOD1G93A mice (Fig. 5H,I; Supplemental Fig. S10A,B). These findings indicated that the absence of ASK1 mitigates motor neuron death in ALS mice.
Discussion
A novel pathogenic mechanism of SOD1mut-mediated ALS is supported by the findings that SOD1mut interacts specifically with Derlin-1 and that this interaction impairs ERAD and leads to ER stress-dependent activation of ASK1 (Fig. 6). The nearly complete loss of SOD1mut-induced ER stress and activation of ASK1 by reduced expression of Derlin-1 or by overexpression of Derlin-1(CT4) strongly suggests that the binding between SOD1mut and Derlin-1 is an important step in the SOD1mut-induced ER stress signaling pathway. It has been reported that translocon-associated protein (TRAP) δ interacts with SOD1mut (Kunst et al. 1997) and that TRAP complex including TRAPδ is required for ERAD (Nagasawa et al. 2007), suggesting that the association of SOD1mut with TRAPδ may also be involved in SOD1mut-induced ER stress.
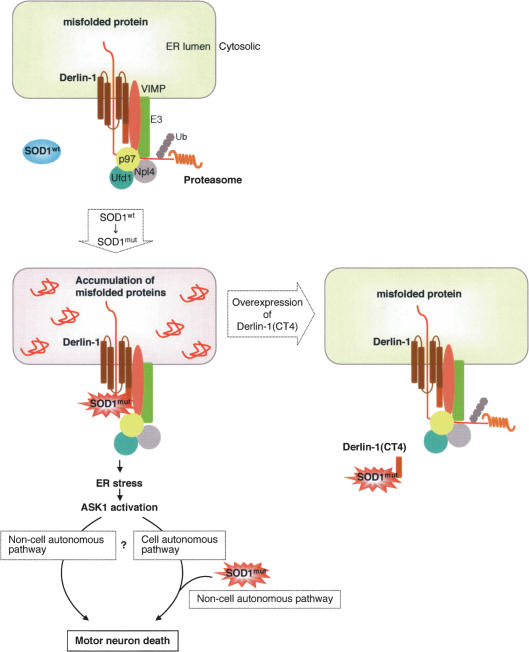
Figure 6.
Schematic representation of the mechanism of SOD1mut-induced motor neuron death through ER stress. See the text for details.
It is unclear how various SOD1mut proteins interact with Derlin-1. SOD1L126Z, a simple C-terminal deletion mutant (Supplemental Fig. S11A), which causes motor neuron disease in mice (Wang et al. 2005), also interacted with Derlin-1 (Supplemental Fig. S11B). Therefore, the Derlin-1 interaction domain of SOD1wt might be concealed by proper folding and be bared by mutation-dependent unfolding of SOD1mut. Another hypothesis is that chaperon proteins might mediate the interaction between SOD1mut and Derlin-1. Although the results from our in vitro studies using recombinant SOD1 proteins support a direct interaction between SOD1mut and Derlin-1 (Fig. 2A), we cannot rule out the possibility that chaperon proteins in the reticulocyte lysate mediate the interaction between SOD1mut and Derlin-1. SOD1mut, but not SOD1wt, is known to interact with heat shock proteins, including Hsp25 and Hsp/Hsc70 (Shinder et al. 2001; Wang et al. 2003; Urushitani et al. 2004).
It is still unclear how the association of SOD1mut with Derlin-1 inhibits the ERAD pathway. Derlin-1 is one of the components required for retro-translocation of misfolded proteins from the ER lumen to the cytoplasm (Lilley and Ploegh 2004; Ye et al. 2004). Derlin-1-mediated retro-translocation is currently believed to occur as follows (Meusser et al. 2005): (1) The substrate is recognized by ER chaperones or ER membrane receptors and is targeted to a retro-translocon; (2) Derlin-1 recruits a membrane complex containing VIMP, the p97 complex, and an E3 ligase; (3) once the substrate emerges into the cytosol, it is captured by p97 as a result of its AAA-ATPase activity and polyubiquitinated by E3; and (4) subsequently, the polyubiquitinated substrates are degraded by the proteasome. The present finding that inhibition of proteasomal activity was undetectable by SOD1mut at the time at which SOD1mut-induced ER stress was evoked (Fig. 1D; Supplemental Fig. S2A) suggests that it is unlikely that SOD1mut primarily targeted final step 4. If SOD1mut disturbs the initiation step 1 of retro-translocation, SOD1mut may inhibit interaction between the unfolded substrate and Derlin-1. However, binding between NHK and Derlin-1 was clearly observed even in SOD1mut-overexpressing cells (Fig. 3D). Moreover, SOD1mut did not inhibit association of Derlin-1 with VIMP, p97, Derlin family proteins, and HRD1 (Fig. 3A–C), suggesting that SOD1mut may not perturb step 2. However, we cannot rule out the possibility that SOD1mut inhibits additional interactions between Derlin-1 and its partners in the ERAD complex. Since ubiquitination of ERAD substrate was inhibited by SOD1mut (Fig. 3E), SOD1mut may thus disturb the presentation of ERAD substrate to the p97 complex or E3. Although exactly how SOD1mut inhibits the directional flow of ERAD substrates remains to be elucidated, it is imaginable that ERAD substrates trapped on the SOD1mut–Derlin-1–VIMP complex would interfere with further disposal of de novo ERAD substrates like a traffic jam, eventually leading to the accumulation of misfolded proteins within the ER lumen.
Although the prolongation of survival of SOD1G93A/ASK1−/− mice clearly indicated the requirement of ASK1 for disease progression, ASK1 deficiency was not sufficient to attenuate disease onset (Fig. 5G; Supplemental Fig. S9). These findings are supported by a recent study that demonstrated the contribution of caspase-9 to motor neuron death and disease progression but not to disease onset in X-chromosome-linked inhibitor of apoptosis (XIAP; a mammalian inhibitor of caspase-3 and caspase-9) transgenic mice (Inoue et al. 2003). We reported previously that ASK1-induced apoptosis is dependent on caspase-9 activation (Hatai et al. 2000), suggesting that the ASK1–caspase-9 proapoptotic pathway may be crucial only for disease progression. On the other hand, several groups have reported that the p38, but not JNK, pathway is activated in motor neurons of the spinal cord of SOD1G93A mice (Wengenack et al. 2004; Holasek et al. 2005; Veglianese et al. 2006). Thus, the ASK1–p38 and/or ASK1–caspase-9 pathway may specifically contribute to SOD1mut-induced motor neuron death.
Since expression of Derlin-1(CT4) and deletion of ASK1 only partially mitigated SOD1mut-induced motor neuron death in vitro (Fig. 5E; Supplemental Fig. S5B), the ER stress–ASK1 pathway-independent cell death mechanism including proteasomal dysfunction (Urushitani et al. 2002; Puttaparthi et al. 2003) may also exist in the pathogenesis of ALS. This may reflect, in part, the incomplete resistance of motor neuron death by ASK1 deficiency in ALS mice (Fig. 5I; Supplemental Fig. S10B). Furthermore, the effect of loss of ASK1 on cell death was slightly weaker than that of Derlin-1(CT4) (Fig. 5E), and Derlin-1(CT4) slightly, although not significantly, enhanced the inhibition of SOD1mut-induced cell death in ASK1−/− neurons (Supplemental Fig. S7). These results suggest that other ER stress-induced proapoptotic pathways, including PERK (Jordan et al. 2002) and CHOP (Zinszner et al. 1998), might also be involved in the downstream SOD1mut-dependent motor neuron death.
Another important issue to be elucidated is the relationship between the ER stress-mediated signaling pathway and the non-cell-autonomous mechanisms of SOD1mut-induced neurotoxicity. Recent studies have emphasized the importance of non-cell-autonomous mechanisms (Clement et al. 2003; Boillee et al. 2006; Yamanaka et al. 2008). Consistent with their findings, SOD1mut induced motor neuron death in spinal cord cultures (Fig. 5C,E; Supplemental Fig. S5A,B) but not in NSC34 cells (data not shown) in which ER stress-mediated ASK1 activation was clearly observed. However, since expression of SOD1mut in neurons alone (Pramatarova et al. 2001) or glial cells alone (Gong et al. 2000) does not induce motor neuron disease in mice, not only non-cell-autonomous mechanisms but also cell-autonomous mechanisms must play roles in SOD1mut-induced neurotoxicity. It is likely that the non-cell-autonomous signals that are transferred from astrocytes, glial cells, or feeder cells to motor neurons are coordinated with and crucial for the ER stress/ASK1-dependent cell-autonomous death signaling induced by SOD1mut (Fig. 6). It is also possible that the ER stress-induced ASK1 activation itself serves as the non-cell-autonomous death signaling (Fig. 6).
In conclusion, our findings demonstrated a novel mechanism by which SOD1mut causes motor neuron death through interaction with Derlin-1, ERAD dysfunction, ER stress, and ASK1 activation. Although further investigation is needed to clarify the mechanisms by which SOD1mut induces the malfunction of Derlin-1 and to identify the downstream effectors of ASK1, Derlin-1 and ASK1 may be potential targets in the treatment of ALS.
Materials and methods
Band-shift analysis for IRE1 and PERK
NSC34 cells were lysed in the lysis buffer as described (Nishitoh et al. 2002). Cell extracts were clarified by centrifugation, and the supernatants were immunoprecipitated with antibodies to IRE1α and PERK. Proteins were resolved by SDS-PAGE under reducing conditions and immunoblotted with antibodies to IRE1α and PERK.
RT–PCR
Total RNA was isolated from 6 × 105 NSC34 cells using ISOGEN kit (Nippongene). Ten micrograms of RNA were reverse-transcribed with SuperScript II (Life Technologies) according to the manufacturer’s instructions. The primers used for PCR were as follows: mouse BiP, 5′-AAGGTCTATGAAGGTGAACGAC CCC-3′ and 5′-GACCCCAAGACATGTGAGCAACTGC-3′; mouse CHOP, 5′-ACTACTCTTGACCCTGCGTCCCTAG-3′ and 5′-CATGTGCAGTGCAGTGCAGGGTCAC-3′; mouse Xbp-1, 5′-GAACCAGGAGTTAAGAACACG-3′ and 5′-AGGCAACA GTGTCAGAGTCC-3′; and mouse G3PDH, 5′-ATGGTGAAG GTCGGTGTGAA-3′ and 5′-ACATGGCCTCCAAGGAGTAA-3′.
Pulse-chase assay
NSC cells were labeled with 35S-Promix (GE Healthcare) in medium lacking methionine and cystein for 30 min, and chased in medium containing excess methionine and cystein. Cells were lysed in a buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 8.0), and protease inhibitor cocktail. An antibody to α1AT was used for immunoprecipitation. Immunoprecipitated samples were resolved by SDS-PAGE and analyzed by an image analyzer.
In vitro binding assay
His6-SOD1wt and His6-SOD1G93A were constructed in pTrcHis vector (Invitrogen). Recombinant His-SOD1wt and His-SOD1G93A proteins were purified with Ni-NTA (Qiagen) according to the manufacturer’s instructions. In vitro translated 35S-labeled VCP, Ufd1, Npl4, VIMP, and Derlin-1 were prepared with the TNT Reticulocyte Lysate System (Promega). 35S-labeled proteins were incubated for 16 h at 4°C with each His-SOD1WT or His-SOD1G93A in buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), and 0.5 mM Triton X-100; washed twice with the washing buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), 0.5% Triton X-100, and 20 mM Imidazole; and analyzed by SDS-PAGE with an image analyzer.
In vivo binding assay
Binding assay using transfected HEK293 cells has been described (Nishitoh et al. 2002). For binding assay between overexpressed Flag-SOD1 and endogenous Derlin-1, ∼5 × 106 NSC34 cells infected with Ad-SOD1wt, Ad-SOD1A4V, Ad-SOD1G85R, or Ad-SOD1G93A for 48 h were immunoprecipitated with an antibody to Flag. For endogenous binding assay between SOD1 and Derlin-1, lysates from tissues of SOD1wt or SOD1G93A mice were immunoprecipitated with an antibody to SOD1 or control nonimmune rabbit polyclonal antibody. For endogenous binding assay between TRAF2 and ASK1, lysates from ∼3 × 107 NSC34 cells were immunoprecipitated with an antibody to TRAF2 or control nonimmune rabbit polyclonal antibody. Proteins were resolved by SDS-PAGE and immunoblotted with antibodies to Derlin-1, SOD1, ASK1, or TRAF2. Aliquots of the same lysates were subjected to immunoblotting with antibodies to Derlin-1 or Flag. For the complex formation assay of IRE1–TRAF2–ASK1, ∼1 × 107 NSC34 cells were infected with adenoviruses and lentivirus for 48 h, and lysates were analyzed by immunoprecipitation-immunoblot (IP-IB) as described (Nishitoh et al. 2002).
Subcellular fractionation
A 30-wk-old mouse spinal cord was washed with PBS at 4°C and placed in ice-cold homogenization buffer (10 mM Tris-HCl at pH 7.4, 1 mM EDTA, protease inhibitors). Tissue was homogenized with 40 strokes of a glass-pestle homogenizer on ice. Homogenates were centrifuged at 14,000g for 7 min. Supernatant was centrifuged at 100,000g for 1 h to yield a microsomal pellet. Microsomes were incubated with homogenization buffer containing 100 mM Na2CO3 (pH 7.5 or pH 10.9) for 5 min, pelleted by centrifugation at 100,000g for 30 min, and analyzed by IB with an antibody to SOD1, Derlin-1, or p97.
In vitro kinase assay
In vitro kinase assay has been described (Nishitoh et al. 2002). In brief, infected 1 × 107 NSC34 cells were lysed with the lysis buffer and immunoprecipitated with an antibody to ASK1. Kinase activity of ASK1 was measured using [γ-32P]ATP and GST-MKK6KN or GST-SEK1KN as substrate and analyzed by SDS-PAGE with an image analyzer.
Ubiquitination assay
Lysates from transfected HEK293 cells were immunoprecipitated with antibody to Flag. After washing with the washing buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), and 5 mM EGTA, beads were boiled with the denaturing buffer containing 1% SDS, 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), and 5 mM EGTA. The supernatant was diluted and reimmunoprecipitated with antibody to Flag and analyzed by SDS-PAGE.
siRNA knockdown of Derlin-1
NSC34 cells were transfected with Derlin-1-specific RNAi oligo and control RNAi oligo (Invitrogen) using Lipofectamine RNAiMAX reagent (Invitrogen). After 24 h, cells were infected with adenovirus for 48 h. Knockdown was analyzed by IB with antibody to Derlin-1. Sequences were as follows: Derl1-MSS228692 (si1) Stealth Select RNAi, AUAUAGUUGAAUC CAAGGAUAACCC and GGGUUAUCCUUGGAUUCAACU AUAU; Derl1-MSS228693 (si2) Stealth Select RNAi, AGUAA CAGGCCUUAAAUCGCGUUCC and GGAACGCGAUUUAA GGCCUGUUACU; and Stealth RNAi Negative Control Medium GC Duplex.
Motor neuron death assay
Spinal cords (Urushitani et al. 2002) were subjected to primary culture for 7 d and infected with lentivirus for 72 h. Cells were stained with antibody to GFAP (1:100; DAKO) and SMI32 antibody (1:1000; Covance).
Mice
Both SOD1G93A transgenic mice (G1L/+ line, backcrossed to C57BL/6 mice; Jackson Laboratories), which expressed human SOD1G93A gene, and SOD1wt transgenic mice, which expressed >8 copies of human SOD1wt gene, were used. An ASK1−/− mouse (Tobiume et al. 2001) was mated with a SOD1G93A mouse under pathogen-free conditions. To circumvent potential difficulties caused by sex of mice, male mice were utilized in the present study. The onset of disease was determined by motor function deficit seen in rota-rod performance at an accelerated speed to 40 rpm for 5 min. All mouse experiments accorded with protocols approved by the Animal Research Committees of Tokyo Medical and Dental University and the University of Tokyo.
Nissl staining
Mice were perfused with PBS followed by 4% PFA in PBS. The spinal cords were excised, fixed with 4% PFA in PBS for 1 d, embedded in paraffin, and sectioned (4 μm). After being deparaffinized using standard protocols, paraffin sections were nissl-stained with cresyl violet. All motor neuron counts were performed in a blinded fashion. We counted the numbers of motor neurons in every fifth section of L3, L4, and L5. Only the large-size neurons with a clear nucleolus and distinctly labeled cytoplasm were included in cell counts.
Acknowledgments
We thank R. Takahashi and M. Urushitani for kind instruction for the primary motor neuron culture; D. Ron, F. Urano, T.A. Rapoport, and Y. Ye for providing antibodies; K. Nagata and N. Hosokawa for providing NHK plasmid; and D. Trono for providing the lentiviral vector system. We also thank all the members of Cell Signaling Laboratory for their critical comments. This study was supported by Grant-in-Aid for Scientific Research on Priority Areas, Research on Pathomechanisms of Brain Disorders, from the Ministry of Education, Culture, Sports, Science and Technology of Japan, CREST, Japan Science and Technology Corporation, Takeda Science Foundation, and The Nakabayashi Trust for ALS Research.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1640108.
References
- Atkin J.D., Farg M.A., Turner B.J., Tomas D., Lysaght J.A., Nunan J., Rembach A., Nagley P., Beart P.M., Cheema S.S., et al. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein disulfide isomerase with superoxide dismutase 1. J. Biol. Chem. 2006;281:30152–30165. doi: 10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- Boillee S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Bowling A.C., Schulz J.B., Brown R.H., Beal M.F. Superoxide dismutase activity, oxidative damage, and mitochondrial energy metabolism in familial and sporadic amyotrophic lateral sclerosis. J. Neurochem. 1993;61:2322–2325. doi: 10.1111/j.1471-4159.1993.tb07478.x. [DOI] [PubMed] [Google Scholar]
- Clement A.M., Nguyen M.D., Roberts E.A., Garcia M.L., Boillee S., Rule M., McMahon A.P., Doucette W., Siwek D., Ferrante R.J., et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- Epstein C.J., Avraham K.B., Lovett M., Smith S., Elroy-Stein O., Rotman G., Bry C., Groner Y. Transgenic mice with increased Cu/Zn-superoxide dismutase activity: Animal model of dosage effects in Down syndrome. Proc. Natl. Acad. Sci. 1987;84:8044–8048. doi: 10.1073/pnas.84.22.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J.P., Tiwari S., Weissman A.M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.H., Parsadanian A.S., Andreeva A., Snider W.D., Elliott J.L. Restricted expression of G86R Cu/Zn superoxide dismutase in astrocytes results in astrocytosis but does not cause motoneuron degeneration. J. Neurosci. 2000;20:660–665. doi: 10.1523/JNEUROSCI.20-02-00660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hatai T., Matsuzawa A., Inoshita S., Mochida Y., Kuroda T., Sakamaki K., Kuida K., Yonehara S., Ichijo H., Takeda K. Execution of apoptosis signal-regulating kinase 1 (ASK1)-induced apoptosis by the mitochondriadependent caspase activation. J. Biol. Chem. 2000;275:26576–26581. doi: 10.1074/jbc.M003412200. [DOI] [PubMed] [Google Scholar]
- Holasek S.S., Wengenack T.M., Kandimalla K.K., Montano C., Gregor D.M., Curran G.L., Poduslo J.F. Activation of the stress-activated MAP kinase, p38, but not JNK in cortical motor neurons during early presymptomatic stages of amyotrophic lateral sclerosis in transgenic mice. Brain Res. 2005;1045:185–198. doi: 10.1016/j.brainres.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Inoue H., Tsukita K., Iwasato T., Suzuki Y., Tomioka M., Tateno M., Nagao M., Kawata A., Saido T.C., Miura M., et al. The crucial role of caspase-9 in the disease progression of a transgenic ALS mouse model. EMBO J. 2003;22:6665–6674. doi: 10.1093/emboj/cdg634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan R., Wang L., Graczyk T.M., Block T.M., Romano P.R. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 2002;76:9588–9599. doi: 10.1128/JVI.76.19.9588-9599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.P., Beaulieu J.M. Cytoskeletal abnormalities in amyotrophic lateral sclerosis: Beneficial or detrimental effects? J. Neurol. Sci. 2000;180:7–14. doi: 10.1016/s0022-510x(00)00422-6. [DOI] [PubMed] [Google Scholar]
- Kikuchi H., Almer G., Yamashita S., Guegan C., Nagai M., Xu Z., Sosunov A.A., McKhann G.M.I., Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl. Acad. Sci. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J., Halligan E., Baptista M.J., Allen S., Heath P.R., Holden H., Barber S.C., Loynes C.A., Wood-Allum C.A., Lunec J., et al. Mutant SOD1 alters the motor neuronal transcriptome: Implications for familial ALS. Brain. 2005;128:1686–1706. doi: 10.1093/brain/awh503. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. ER quality control: The cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Kunst C.B., Mezey E., Brownstein M.J., Patterson D. Mutations in SOD1 associated with amyotrophic lateral sclerosis cause novel protein interactions. Nat. Genet. 1997;15:91–94. doi: 10.1038/ng0197-91. [DOI] [PubMed] [Google Scholar]
- Li M., Ona V.O., Guegan C., Chen M., Jackson-Lewis V., Andrews L.J., Olszewski A.J., Stieg P.E., Lee J.P., Przedborski S., et al. Functional role of caspase-1 and caspase-3 in an ALS transgenic mouse model. Science. 2000;288:335–339. doi: 10.1126/science.288.5464.335. [DOI] [PubMed] [Google Scholar]
- Lilley B.N., Ploegh H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Lilley B.N., Ploegh H.L. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lillo C., Jonsson P.A., Vande Velde C., Ward C.M., Miller T.M., Subramaniam J.R., Rothstein J.D., Marklund S., Andersen P.M., et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: The long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Nagasawa K., Higashi T., Hosokawa N., Kaufman R.J., Nagata K. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep. 2007;8:483–489. doi: 10.1038/sj.embor.7400933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Okada T., Yoshida H., Kaufman R.J., Nagata K., Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P., Borchelt D.R., Houseweart M.K., Cleveland D.W., Brown R.H. Caspase-1 is activated in neural cells and tissue with amyotrophic lateral sclerosis-associated mutations in copper–zinc superoxide dismutase. Proc. Natl. Acad. Sci. 1998;95:15763–15768. doi: 10.1073/pnas.95.26.15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramatarova A., Laganiere J., Roussel J., Brisebois K., Rouleau G.A. Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J. Neurosci. 2001;21:3369–3374. doi: 10.1523/JNEUROSCI.21-10-03369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaparthi K., Wojcik C., Rajendran B., DeMartino G.N., Elliott J.L. Aggregate formation in the spinal cord of mutant SOD1 transgenic mice is reversible and mediated by proteasomes. J. Neurochem. 2003;87:851–860. doi: 10.1046/j.1471-4159.2003.02028.x. [DOI] [PubMed] [Google Scholar]
- Raoul C., Estevez A.G., Nishimune H., Cleveland D.W., deLapeyriere O., Henderson C.E., Haase G., Pettmann B. Motoneuron death triggered by a specific pathway downstream of Fas. potentiation by ALS-linked SOD1 mutations. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P.M., Seeger M. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Takeda K., Ichijo H. The ASK1–MAP kinase signaling in ER stress and neurodegenerative diseases. Curr. Mol. Med. 2006;6:87–97. doi: 10.2174/156652406775574541. [DOI] [PubMed] [Google Scholar]
- Shinder G.A., Lacourse M.C., Minotti S., Durham H.D. Mutant Cu/Zn-superoxide dismutase proteins have altered solubility and interact with heat shock/stress proteins in models of amyotrophic lateral sclerosis. J. Biol. Chem. 2001;276:12791–12796. doi: 10.1074/jbc.M010759200. [DOI] [PubMed] [Google Scholar]
- Sifers R.N., Brashears-Macatee S., Kidd V.J., Muensch H., Woo S.L. A frameshift mutation results in a truncated α 1-antitrypsin that is retained within the rough endoplasmic reticulum. J. Biol. Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Suzuki T., Park H., Lennarz W.J. Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: Occurrence, primary structure, and potential functions. FASEB J. 2002;16:635–641. doi: 10.1096/fj.01-0889rev. [DOI] [PubMed] [Google Scholar]
- Tobisawa S., Hozumi Y., Arawaka S., Koyama S., Wada M., Nagai M., Aoki M., Itoyama Y., Goto K., Kato T. Mutant SOD1 linked to familial amyotrophic lateral sclerosis, but not wild-type SOD1, induces ER stress in COS7 cells and transgenic mice. Biochem. Biophys. Res. Commun. 2003;303:496–503. doi: 10.1016/s0006-291x(03)00353-x. [DOI] [PubMed] [Google Scholar]
- Tobiume K., Matsuzawa A., Takahashi T., Nishitoh H., Morita K., Takeda K., Minowa O., Miyazono K., Noda T., Ichijo H. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2:222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T.A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Urushitani M., Kurisu J., Tsukita K., Takahashi R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. J. Neurochem. 2002;83:1030–1042. doi: 10.1046/j.1471-4159.2002.01211.x. [DOI] [PubMed] [Google Scholar]
- Urushitani M., Kurisu J., Tateno M., Hatakeyama S., Nakayama K., Kato S., Takahashi R. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J. Neurochem. 2004;90:231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J.P. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat. Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- Veglianese P., Lo Coco D., Bao Cutrona M., Magnoni R., Pennacchini D., Pozzi B., Gowing G., Julien J.P., Tortarolo M., Bendotti C. Activation of the p38MAPK cascade is associated with upregulation of TNF α receptors in the spinal motor neurons of mouse models of familial ALS. Mol. Cell. Neurosci. 2006;31:218–231. doi: 10.1016/j.mcn.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N.G., Jenkins N.A., Borchelt D.R. Copper-binding-site-null SOD1 causes ALS in transgenic mice: Aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- Wang J., Xu G., Li H., Gonzales V., Fromholt D., Karch C., Copeland N.G., Jenkins N.A., Borchelt D.R. Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: αB-crystallin modulates aggregation. Hum. Mol. Genet. 2005;14:2335–2347. doi: 10.1093/hmg/ddi236. [DOI] [PubMed] [Google Scholar]
- Wengenack T.M., Holasek S.S., Montano C.M., Gregor D., Curran G.L., Poduslo J.F. Activation of programmed cell death markers in ventral horn motor neurons during early presymptomatic stages of amyotrophic lateral sclerosis in a transgenic mouse model. Brain Res. 2004;1027:73–86. doi: 10.1016/j.brainres.2004.08.054. [DOI] [PubMed] [Google Scholar]
- Wong P.C., Pardo C.A., Borchelt D.R., Lee M.K., Copeland N.G., Jenkins N.A., Sisodia S.S., Cleveland D.W., Price D.L. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T.A. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T., Ishigaki S., Yamamoto M., Liang Y., Niwa J., Takeuchi H., Doyu M., Sobue G. Differential expression of inflammation- and apoptosis-related genes in spinal cords of a mutant SOD1 transgenic mouse model of familial amyotrophic lateral sclerosis. J. Neurochem. 2002;80:158–167. doi: 10.1046/j.0022-3042.2001.00683.x. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes & Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]