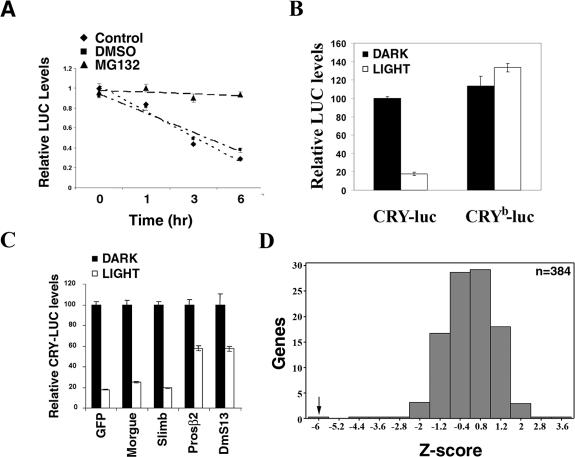
Figure 1.
Development of a primary screen to identify novel components involved in light-dependent CRY degradation. (A) Light-dependent degradation of a CRY-luc fusion protein: S2R+ cells were transfected with expression constructs for a Cry-luc fusion protein and Actin-Renilla luciferase. Cells were exposed to light in the presence of DMSO (control) or proteasome inhibitor MG132 (100 μM) for the indicated times, and luciferase activity was monitored. Luciferase activity was normalized to that of cells kept in darkness, set as 1. Average values from three samples are plotted, and the error bars depict standard error of the mean (SEM). The results are representative of two independent experiments. Light-dependent degradation of the CRY-luciferase fusion protein was blocked by proteasome inhibitor MG132. (B) A single point mutation in the flavin-binding pocket of CRY, CRYb, renders it insensitive to light. S2R+ cells were transfected with expression constructs for CRY-luc or CRYb-luc fusion protein in 384-well plates and assayed for luciferase activity after light exposure for 6 h. Luciferase activity from three samples is plotted relative to the CRY activity in transfected cells kept in the dark (set to 100%); error bars represent SEM. In response to light exposure, activity of the CRY-luc fusion protein was reduced by 83% (P-value = 2 × 10−9), while activity of the CRYb-luc fusion protein was unchanged. Similar results were obtained in three experiments with different cell densities. (C) RNAi of 26S proteasome components blocks light-dependent CRY degradation. S2R+ cells were cotransfected with the CRY-luc expression construct and dsRNAs, as indicated, in 384-well plates. Act-Renilla luc was included as a transfection control. Ninety-six hours post-transfection, cells were exposed to light and luciferase activity was measured as indicated above. Relative luciferase activity from three samples is plotted; error bars represent SEM. The value obtained with dsRNA against gfp in the dark was set as 100% and used for normalization. dsRNA against cry, which dramatically reduced luciferase activity (data not shown), was used as a positive control. RNAi of the 26S proteasome core components Prosβ2 and DmS13 (also called as Rpn11) significantly blocked light-dependent CRY-luc degradation. Similar results were obtained in two independent experiments. (D) A pilot screen demonstrates the validity of the CRY-luc assay. The light-dependent CRY-luc degradation assay was performed in S2R+ cells with 380 randomly picked dsRNAs from the genomic collection. dsRNA against the proteasome component Prosβ2 was used as a positive control; thread dsRNA was included as a control to verify the efficiency of RNAi. Normalized luciferase levels were compared between the dark- and light-treated plates. Z-scores, standard deviations from the plate mean (see the Materials and Methods for details), are plotted. While dsRNA against Prosβ2 significantly blocked light-dependent CRY-luc degradation (Z-score = −6.2), most dsRNAs had no effect. Similar results were obtained in two independent experiments.