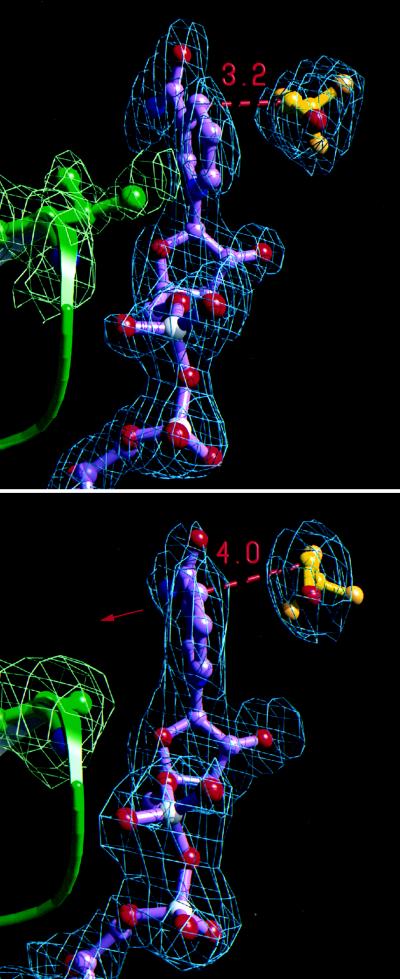
Figure 2.
Comparison of active site structures from a high tunneling (Phe-93 → Trp, top) and a low tunneling (Val-203 → Ala, bottom) mutant of LADH. Electron density omit maps (Fo − Fc) and resulting models for the residue at position 203 (green), NAD+ (C, purple; O, red; P, white; N, blue), and trifluoroethanol (C, yellow; F, orange; O, red) are illustrated for each structure. Omit maps were generated with trifluoroethanol, NAD+, and residue-203 omitted from the final model and are contoured at σ levels of 2.5 (top) and 2.0 (bottom). (Upper): The nicotinamide ring in Phe-93 → Trp is in van der Waals contact with a methyl group of Val-203. The average donor to acceptor carbon distance among the two independent monomers is 3.2 Å. (Lower): In Val-203 → Ala, van der Waals contact between residue 203 and the nicotinamide ring is removed, causing a shift in ring position (red arrow, see Fig. 3). The average donor to acceptor carbon distance among the four crystallographically independent monomers is 4.0 Å. [Figs. 2 and 4 were rendered using molscript (24), rayscript (E. Fontana, D. Peisach, and E. Peisach, Brandeis University) and rayshade (version 4.0, C. Kolb and R. Bogart, Princeton University).]