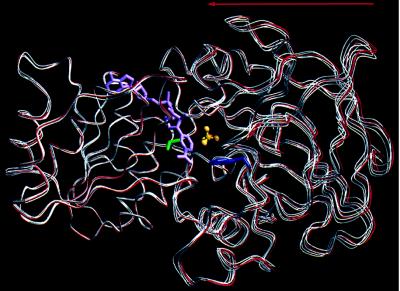
Figure 4.
Difference in interdomain conformation between the hyperclosed Val-203 → Ala LADH structure and the closed Phe-93 → Trp structure. Cofactor domains at left were aligned by a least-squares fit of the two crystallographically independent Phe-93 → Trp monomers (red) and the four independent Val-203 → Ala monomers (white). The hyperclosed conformation of the Val-203 → Ala mutant results from an ≈0.5-Å average rigid body translation of its catalytic domain toward its coenzyme domain, when compared with the closed conformation of the Phe-93 → Trp structure. Vectors describing the rigid body translation for each Val-203 → Ala monomer correlate well, all lying within 25° of the average translation vector (arrow). Rotational components of this motion range between 0.5 and 2.5°, but there is little directional correlation between rotation axes for each of the four independent monomers. Analysis of interdomain motion used CCP4 least-squares fitting routine Lsqkab (32, 33). Val-203 (green), NAD+ (purple), Trp-93 (blue), and trifluoroethanol (C, yellow; F, orange; O, red) are displayed from the Phe-93 → Trp structure.