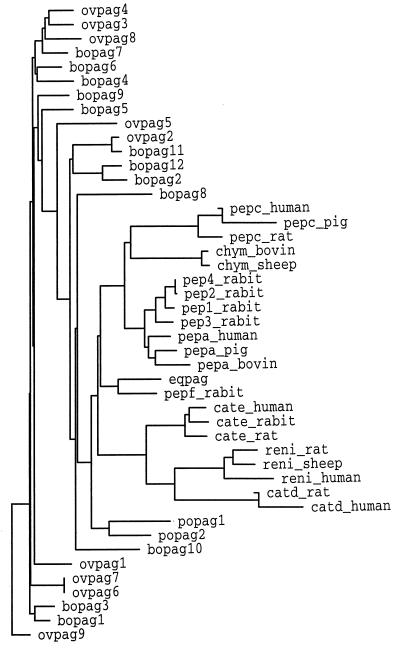
Figure 8.
A growtree phylogram based on amino acid sequences showing the relationship of all known cloned PAGs to the commoner mammalian aspartic proteinases whose sequences are known. The tree was constructed by the Wisconsin GCG programs distances and growtree. The lengths of the branches are proportional to the degree of amino acid diversity within pairs of proteins. Occasional negative branch lengths result because the neighbor-joining algorithm always attempts to create an additive tree, even when this is not possible. Analyses were carried out over the first 220 amino acids so as to include PAG sequences with missing exons. Protein data bank symbols are as follows: pep, pepsin; chym, chymosin; cate, cathepsin E, catd, cathepsin D; reni, renin; rabit, rabbit; bovin, bovine.