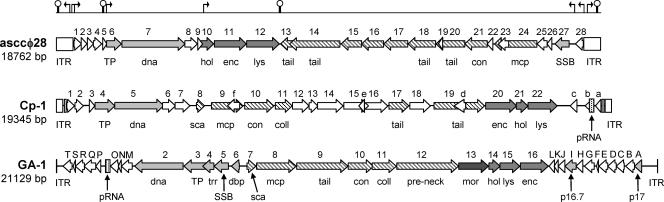
FIG. 4.
Deduced genetic map of phage asccφ28 compared with maps for phages Cp-1 (29) and GA-1, a φ29-like phage (30). The maps for Cp-1 and GA-1 are derived from data in the GenBank database (accession numbers Z47794 and X96987, respectively). Gene functions are abbreviated as follows: TP, terminal protein; dna, DNA polymerase; hol, holin; enc, encapsidation protein; lys, lysin; tail, tail protein; coll, collar protein; mcp, major capsid (head) protein; con, collar or connector protein; SSB, single-stranded DNA-binding protein; sca, scaffolding protein; trr, transcriptional regulator; dbp, DNA-binding protein; pre-neck, preneck appendage protein; mor, morphogenesis protein. The genes encoding DNA replication proteins p16.7 and p17 in GA-1 are indicated. Light gray shading indicates genes involved in DNA replication or transcriptional regulation; medium gray shading indicates genes involved in cell lysis; diagonal lines indicate genes encoding structural proteins; and dark gray shading in the GA-1 map indicates the gene encoding the morphogenesis protein. Proteins with no known function are indicated by open arrows. Packaging RNAs and inverted terminal repeat regions are indicated by pRNA and ITR, respectively. Above the asccφ28 map, putative promoters and rho-independent transcriptional terminators (see the GenBank accession number EU438902 sequence for details) are indicated by arrows and hairpin structures, respectively.