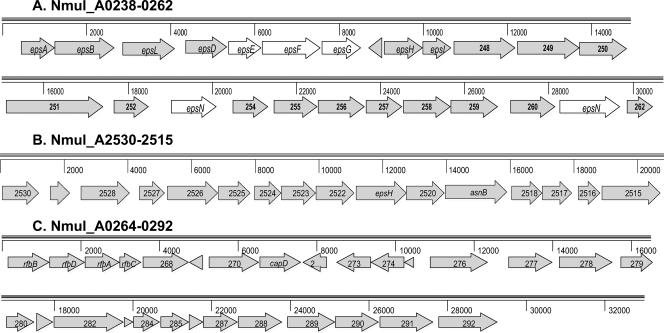
FIG. 3.
EPS, LPS, and capsule clusters in N. multiformis. (A) Nmul_A0238 to Nmul_A0262 (Nmul_A0238-0262). EPS cluster with similarity to cluster in Methylobacillus sp. strain 12S (AB062506). Gene designations for orthologs in Methylobacillus sp. strain 12S are given, and genes essential for the production of the EPS methanolan in Methylobacillus sp. strain 12S are highlighted. (B) Putative EPS and LPS cluster including Nmul_A2530 to Nmul_A2515 (Nmul_A2530-2515) and their putative products and functions are as follows: Nmul_A2529, polysaccharide export to OM protein; Nmul_A2528, EPS biosynthesis chain length determinant similar to Wzz; Nmul_A2527, capsular polysaccharide biosynthesis protein; Nmul_A2526, signal peptide motif for secreted protein of unknown function; Nmul_A2525, type II transport ATPase; Nmul_A2524, polysaccharide deacetylase; Nmul_A2523, conserved hypothetical protein; Nmul_A2522, glycosyltransferase; Nmul_A2521 (epsH), EpsH (EPS locus protein H) involved in processing proteins to OM locations; Nmul_A2520, glycosyltransferase group 1, similar to wapH in P. putida (PP_4943); Nmul_A2519, asparagine synthase; Nmul_A2516, methyltransferase; Nmul_A2515, FkbH domain membrane protein involved in bacterial cell division. (C) Nmul_A0264-0292 putative EPS and LPS gene clusters. Nmul_A0264 to Nmul_A0292 (Nmul_A0264-0292) putative EPS and LPS gene clusters encoding proteins are shown as follows: Nmul_A0264 to Nmul_A0267, synthesis of dTDP-l-rhamnose (rfbBDAC); Nmul_A0268, fatty acid desaturase; Nmul_A0270, UDP-glucose/GDP-mannose dehydrogenase; Nmul_A0271, polysaccharide biosynthesis CapD; Nmul_A0277, dolichyl-P-beta-d-mannosyl transferase; Nmul_A0278 to Nmul_A0292, several glycosyltransferases, capK gene, and a polysaccharide biosynthesis protein often implicated in polysaccharide capsule biosynthesis.