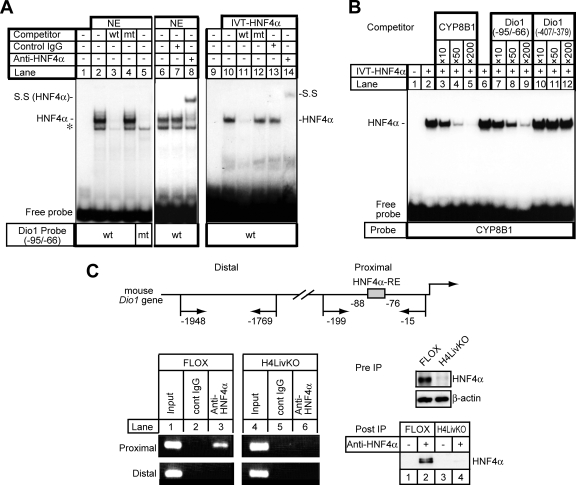
FIG. 3.
Specific binding of HNF4α to the mouse Dio1 gene promoter. (A) EMSA. Nuclear extracts (NE) isolated from HepG2 cells (20 μg) or in vitro-translated HNF4α protein (IVT-HNF4α) were incubated with a 32P-labeled wild-type (wt) (lanes 1 to 4 and 6 to 8) or mutant (mt) (lane 5) oligonucleotide probe carrying HNF4α-RE in the absence or presence of a 200-fold excess of the unlabeled wild-type (lanes 3 and 11) or mutant oligonucleotide (lanes 4 and 12). For the supershift assay, an anti-HNF4α antibody, IgG-H1415 (lanes 8 and 14), was added to the binding reaction mixture. Each anti-HNF4α antibody (2 μg) was added to the reaction mixture 30 min after the addition of the probe and incubated for an additional 10 min. The HNF4α-DNA complex and its supershifted complex (S.S) is indicated. Nonspecific bands are denoted by an asterisk. (B) IVT-HNF4α was incubated with a double-stranded, radiolabeled oligonucleotide probe containing the sequence of the high-affinity HNF4α binding site of the human CYP8B1 promoter (70), followed by EMSA. Competition experiments were performed in the presence of a 10- to 200-fold molar excess of the unlabeled oligonucleotides for the HNF4α binding site of the human CYP8B1 promoter (lanes 3 to 5), the DR1 motif at −88 (Dio1 −95/−66) (lanes 7 to 9), or the DR1 motif at −400 (Dio1 −407/−379) (lanes 10 to 12) of the mouse Dio1 promoter. (C) HNF4α-RE of the mouse Dio1 promoter binds to HNF4α in the context of an intact chromatin structure as demonstrated by ChIP. Schematic representation of the 5′-FR of the mouse Dio1 gene. Positions of primer sets (arrow) relative to the translation start site are denoted (top). Cross-linked DNA-protein complexes from either FLOX or H4LivKO mouse liver extracts were immunoprecipitated with anti-HNF4α and control IgG, followed by PCR amplification with specific primers as schematically depicted in the diagram. Genomic DNA in the input cell lysates was used as a positive control. As a negative control, the distal region, which has no HNF4α binding sites, was used for PCR amplification. Amplified PCR products were separated on an ethidium bromide-stained 2% agarose gel (bottom left). The presence of HNF4α in cell lysate before immunoprecipitation (Pre IP) and the samples immunoprecipitated with anti-HNF4α antibody (Post IP), detected by immunoblot analysis with anti-HNF4α antibody, is shown (bottom right).