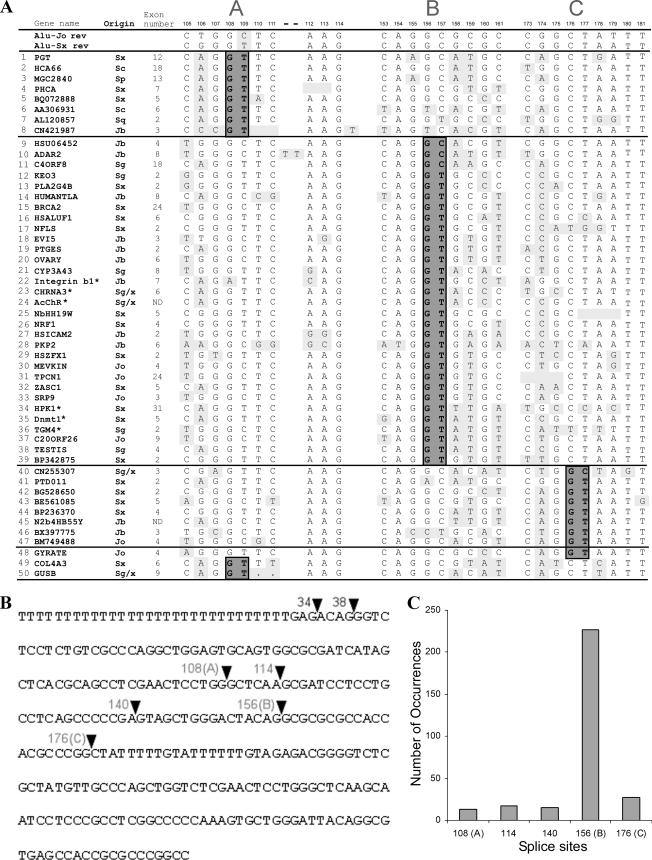
FIG. 1.
5′ss selection of Alu-derived exons. (A) Alignment of 50 exonized Alu elements in the antisense orientation with respect to the pre-mRNA. This data set is based on previous studies (25, 36, 41) as well as on further literature (marked by asterisks; see Table S1 in the supplemental material for references). The 26 nt presented contain three possible 5′ss selected during Alu exonization. The first two intronic positions at each site are highlighted in dark gray and marked 5′ss A, B, and C. Consensus sequences of subfamilies S and Jo appear in the first two rows. Single mutations differing from the ancestral S and Jo subfamilies are highlighted in light gray. Rows 48 to 50 represent the 5′ss of Alu sequences whose constitutive exonization was shown to cause a genetic disease, either OAT deficiency (GYRATE), Alport syndrome (COL4A3), or Sly syndrome (GUSB). The mutation that causes Alport syndrome is in the 3′ss region (−7G to T), as shown previously (25). The mutations causing Sly syndrome and OAT deficiency are both in 5′ss regions: the mutation causing OAT syndrome is in row 48, position 176, and the mutations resulting in Sly deficiency are in row 50, positions 110 and 111. See Table S1 in the supplemental material for references for these three genetic diseases. Gene names are given according to RefSeq conventions, and the Alu exon number is the exon serial number for each gene. (B) Most frequently used splice sites in Alu right-arm exonizations in the antisense orientation, mapped onto the Alu-Jo consensus sequence (19). The 3′ss at positions 34 and 38 and the 5′ss at positions 108, 114, 140, 156, and 176 are indicated with arrowheads marking the exon-intron junctions at these sites. (C) Frequencies of selection of the five main 5′ss resulting in Alu exonization.