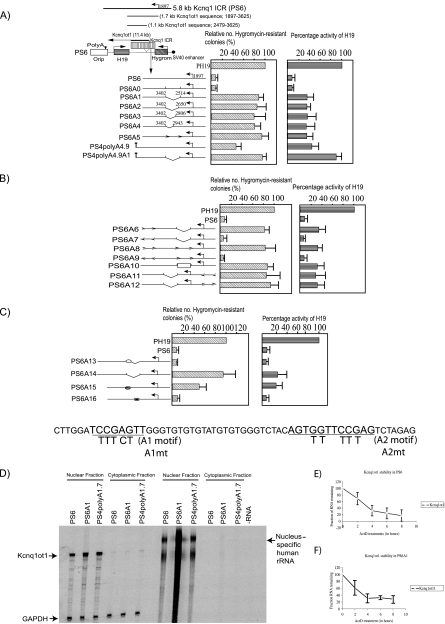
FIG. 2.
The Kcnq1ot1 SD at the 5′ end of the Kcnq1ot1 antisense RNA silences flanking genes in an orientation- and transcription-dependent manner. (A to C) Bar graphs show the activities of the hygromycin and H19 genes, analyzed for each construct. The maps alongside the bar graphs depict the wild-type and mutant forms of the 5.8-kb Kcnq1 ICR, which were inserted into the PH19 episome between the H19 and hygromycin genes. The activities of reporter genes in each construct are shown as percent expression levels relative to those of control vectors (PH19 for the H19 and hygromycin genes). The expression levels of H19 were quantified by RPA 8 days after transient transfection of wild-type or mutant constructs into the JEG-3 cell line. The levels were normalized against total input RNA (using the GAPDH control) and episome copy numbers (by Southern hybridization of genomic DNA from the transfected cells with an episome-specific probe and an internal β-actin control probe). Hygromycin gene activity was analyzed by counting the hygromycin-resistant cell colonies after selection with hygromycin. RPA results for H19 are expressed as means ± standard deviations for three independent experiments; hygromycin-resistant colony counts are expressed as means ± standard deviations for a minimum of six independent experiments. (D) Episome-encoded Kcnq1ot1 is specifically localized in the nuclear compartment, and selective deletion of the silencing domain from Kcnq1ot1 or truncation of Kcnq1ot1 has no effect on its nuclear localization. (E and F) The half-life of the episome-encoded Kcnq1ot1 transcript is 3.2 h (E), and selective deletion of silencing domain has no significant effect on the stability of Kcnq1ot1 (F).