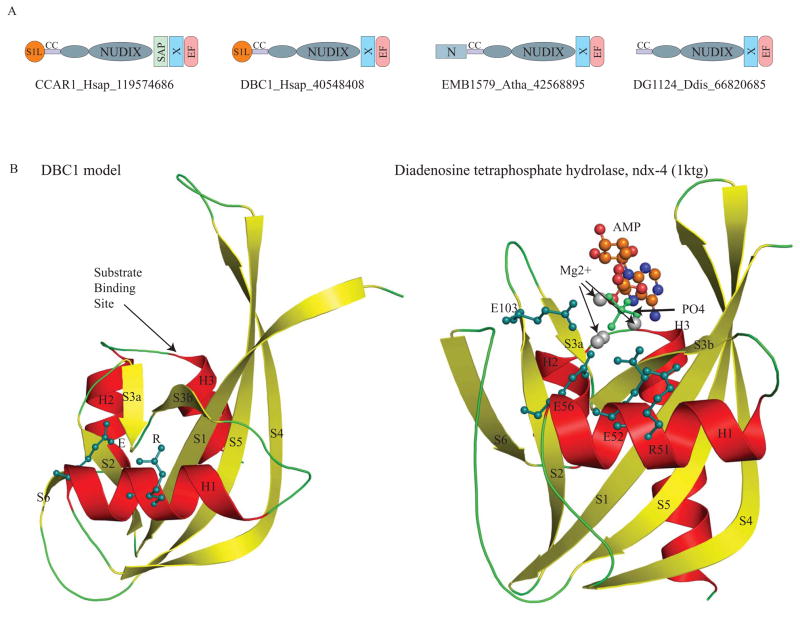
Fig. 2. A. Domain architectures of the DBC1 family.
The domain architectures found in the DBC1 family are shown. Domain architectures are labeled with a representative gene name, the species abbreviation, and the Genebank identifier (GI) number separated by underscores. The N-terminal extension of the Nudix domain is shown as a small ellipse. Domain abbreviations are: S1L – S1-like OB fold domain; EF – EF hand; CC – coiled coil region; and N – N terminal domain specific to plant members of the DBC1 family.
B. Model of the DBC1 Nudix domain compared with the crystal structure of the catalytically active version ndx-4 (pdb: 1ktgA).
The DBC1 model was constructed using DR1025, a nucleotide pyrophosphatase (PDB: 1SU2), DR1184, a CoA pyrophosphatase (PDB: 1NQZ), both from Deinococcus and ndx-4, a diadenosine tetraphosphate hydrolase(PDB: 1KTG) from Caenorhabditis, as templates. The crystal structure of ndx-4, a diadenosine tetraphosphate hydrolase (1KTG), is shown for comparison. The conserved active-site residues, the substrate binding site, Magnesium ions, phosphate ion and the substrate ADP are shown.