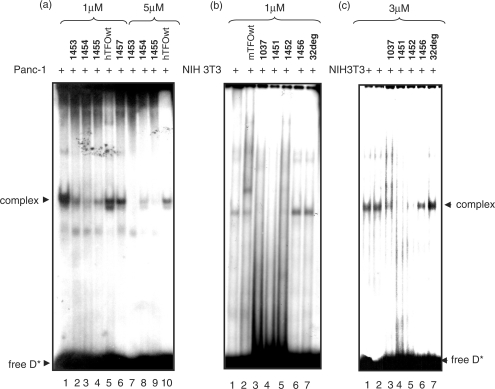
Figure 8.
EMSA showing the abrogation by the designed TINA TFOs of a DNA–protein complex C1 formed between the murine and human KRAS pur/pyr element and a nuclear extract. (a) DNA–protein interaction between a 32-mer human duplex (5′-AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG; 5′-CCTCCCCCTCTTCCCTCTTCCCACACCGCCCT) and Panc-1 nuclear extract in the presence or absence of TFO. Loading: human target (10 nM) with 3 μg extract (lane 1); human target (10 nM) + 1 μM 1453, 1454, 1455, hTFOwt and 1457 incubated overnight at 37°C in binding buffer (see EMSA), then the mixtures treated with 3 μg extract for 2 h (lanes 2–6); human target (10 nM) + 5 μM 1453, 1454, 1455 and hTFOwt incubated overnight, then the mixtures treated for 2 h with 3 μg extract (lanes 7–10); (b) DNA–protein interaction between a 39-mer murine duplex (5′-CGCGCGGGAGGGAGGGAAGGAGGGAGGGAGGGAGCGGCT; 5′-AGCCGCTCCCTCCCTCCCTCCTTCC-CTCC-CTCCCGCGCG) and NIH 3T3 nuclear extract in the presence or absence of TFO. Loading: 39-mer murine duplex (10 nM) incubated with 12 μg NIH 3T3 extract for 2 h (lane 1); 32-mer duplex (10 nM) + 1 μM mTFOwt; 1037, 1451, 1452, 1456, 32deg incubated overnight in binding buffer at 37°C, then treated for 2 h with 12 μg extract (lanes 2–7); (c) 39-mer murine duplex (10 nM) added simultaneously with 12 μg NIH 3T3 extract and 3 μM 1037, 1451, 1452, 1456, 32deg and incubated for 2 h (lanes 3–7); duplex with only 12 μg NIH 3T3 extract (lanes 1 and 2). The sequence of 32deg is: 5′-GCATTCTGATTACACGTATTACCTTCACTCCA.