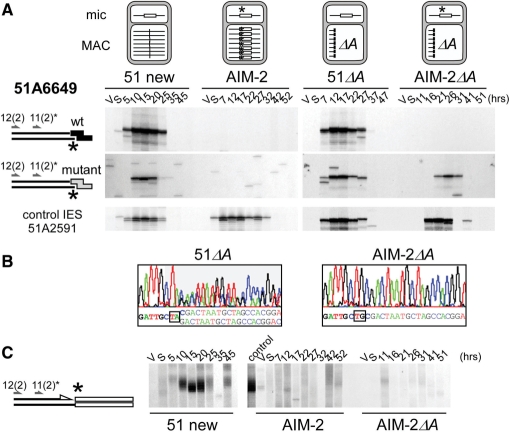
Figure 5.
Effect of a germline mutation at the left end of IES 51A6649 on the cleavage of the mutant end. (A) LMPCR detection of broken chromosome ends in strains 51 new, AIM-2, 51ΔA and AIM-2ΔA, using linkers I′/(CTAC)J′ (wt: top panels) or I′/(CTGC)J′ (mutant: middle panels). The structure of the A locus in the maternal macronucleus of each strain is shown on top of the figure, with IES 51A6649 drawn as a white box and its mutant end marked with an asterisk. Bottom panels: broken chromosome ends at the left end of control IES 51A2591 analyzed as in Figure 3. Autogamy time-points are indicated on top of the figure. (B) En masse sequencing of the re-amplified LMPCR products obtained following ligation of mutant linker I′/(CTAC)J′ to the T17 and T26 genomic DNA samples from 51ΔA and AIM-2ΔA, respectively. Oligonucleotide 51A6649-11(2) was used as a sequencing primer. (C) Terminal transferase-mediated poly(dG) tailing of the free 3′ hydroxyl ends on the macronucleus-destined side of IES 51A6649 left boundary, during autogamy of strains 51 new, AIM-2 and AIM-2ΔA. As a control, the T20 sample from 51 new was loaded next to the AIM-2 samples. Oligonucleotides used for PCR and primer extension are indicated by arrows.