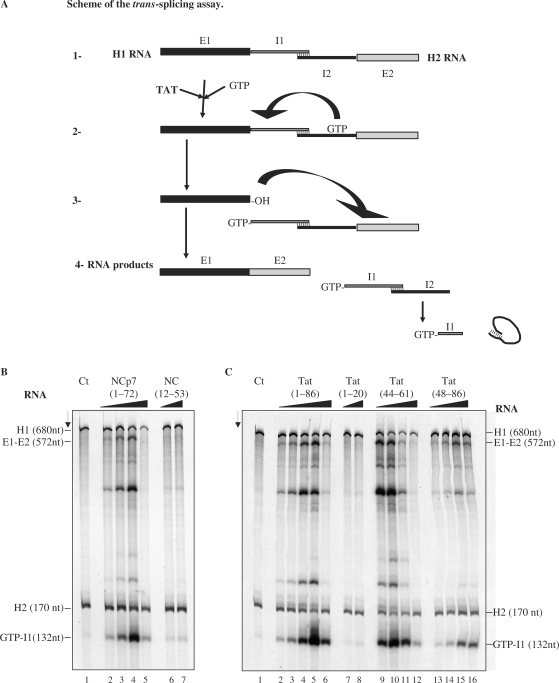
Figure 6.
Enhancement of trans-splicing by HIV-1 Tat. (A) Schematic representation of the trans-splicing assay: The two RNA constructs H1 (encoding exon 1 and the 5′ part of the intron) and H2 (encoding the 3′ part of the intron and exon 2) have to fold into a splicing competent structure (1). The splicing reaction was started by addition of the 32P-labeled GTP. The nucleic acid chaperone was eventually added at step 2. The final RNA products are represented in (4). The splicing rates are based on the levels of GTP-I1 (see B). (B and C). Assays where the H1 and H2 RNAs were incubated with or without a nucleic acid chaperone. At the end of the reaction RNAs were phenol treated to remove the protein and analysed by PAGE in denaturing conditions (see Materials and methods section). The RNA substrates (H1, H2), the ligated exons (E1-E2) and the product (guanosine-5′-intron G-I1) are indicated. Panel B. Lane 1: splicing reaction at 37°C with RNA alone at a concentration of 4 × 10−8 M. Lanes 2–5: HIV-1 NCp7 at concentrations of 2.5, 5, 10 and 20 × 10−7 M, corresponding to protein to nt ratios of 1:128, 1:64, 1:32 and 1:16. Lanes 6 and 7: NC(12–53) at concentrations of 1 and 2 × 10−6 M, corresponding to peptide to nt ratios of 1:32 and 1:16. Panel C. Lane 1: splicing reaction at 37°C with RNA alone at a concentration of 4 × 10−8 M. Lanes 2–6: Tat(1–86) at concentrations of 1.25, 2.5, 5, 10 and 20 × 10−7 M, corresponding to protein to nt molar ratios of 1:256, 1:128, 1:64, 1:32 and 1:16. Lanes 7 and 8: Tat(1–20) at concentrations of 1 and 2 × 10−6 M. Lanes 9–12: Tat(44–61) at concentrations of 2, 4, 8 and 16 × 10−7 M, corresponding to peptide to nt molar ratios of 1:170, 1:85, 1:42.5 and 1:21.2. Lanes 13–16: Tat(48–86) at concentrations of 2, 4, 8 and 16 × 10−7 M, corresponding to molar ratios of 1:170, 1:85, 1:42.5 and 1:21.2. Note that HIV-1 NCp7 and Tat strongly activated trans-splicing at protein to nt molar ratios of 1:32 (compare lanes 1 and 4 in B, and lanes 1 and 5 in C). Tat peptide (44–61) was also very active (lane 10 in C). Peptides NC(12–53) and Tat(1–20) were very poorly active.