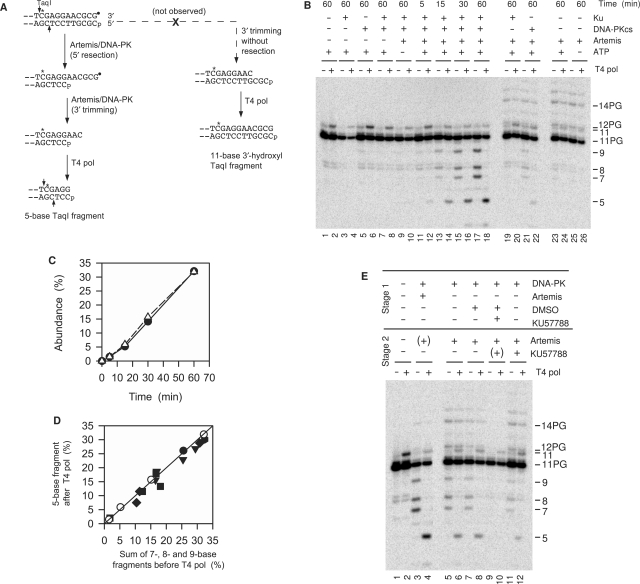
Figure 2.
Coordinate 5′ and 3′ trimming of a blunt 3′-PG (filled circle) 5′-phosphate substrate by Artemis/DNA-PK. (A) Experimental rationale. If 3′ trimming is accompanied by 5′ trimming (left pathway), subsequent treatment with T4 pol will resect or fill in the top strand to match the bottom strand. If 3′ trimming occurs without 5′ trimming (right pathway), T4 pol will fill in the gap, resulting in an increase in labeled 11-base 3′-hydroxyl fragments. (B) Time course and requirements for 3′ and concomitant 5′ trimming. Samples were treated for the indicated times with 25 nM Ku, 50 nM DNA-PKcs and/or 90 nM Artemis. Following deproteinization, half of each sample was treated with T4 pol, and then all samples were treated with TaqI. (C) Time course for accumulation of 3′-trimmed 7-, 8- and 9-base fragments without T4 pol treatment (filled circle) or 5-base fragment with T4 pol treatment (open triangle). (D) Comparison of trimming before and after T4 pol treatment; different symbols indicate six independent experiments similar to that shown in (B). (E) Effect of the DNA-PKcs inhibitor KU57788 on 3′ and 5′ trimming by Artemis in a two-stage reaction. In stage 1, samples were treated with Ku, DNA-PKcs and/or Artemis in the presence or absence of 1 μM KU57788 or 2% DMSO for 30 min. Artemis and/or KU57788 was then added and the sample incubated for an additional 45 min. A ‘(+)’ indicates that a component was present in stage 2 because it had been added in stage 1. Protein concentrations are the same as in (B).