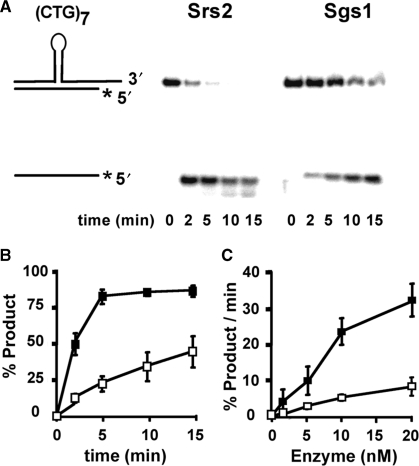
Figure 2.
Time- and enzyme concentration-dependent unwinding of (CTG)7 hairpin-containing duplexes. The schematic diagram shows the structured hairpin embedded within normal duplex regions. The shorter strand is radiolabeled on the 5′ end, as denoted by the asterisk. Purified Srs2 and Sgs1 were incubated with the DNA substrates (0.5 nM) for the indicated times. For 0 min, the substrate was incubated without protein. The reactions were quenched and the products were resolved on 12% nondenaturing polyacrylamide gel and visualized by phosphorimaging. (A) Srs2 or Sgs1 (10 nM each) were incubated with the substrate for 0–15 min. (B) Time dependence of unwinding from two–three repetitions (including that shown in panel A). (C) Enzyme concentration effects on unwinding rates in the linear time range, calculated as percentage of product unwound per minute of reaction. For (B) and (C), error bars are the range of values observed; filled squares, Srs2; unfilled squares, Sgs1.