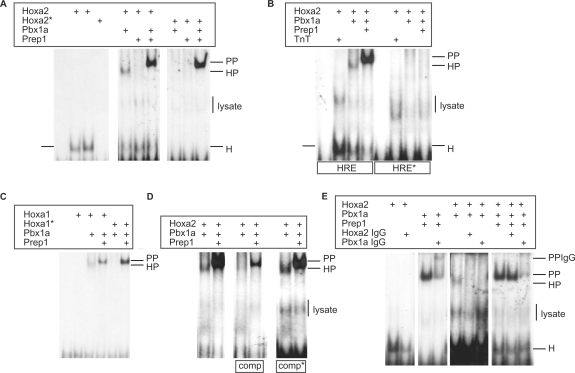
Figure 4.
Hoxa2 and Pbx1a bind to the HRE sequence. (A) Gel retardation assays were performed with double-stranded oligonucleotides bearing the wild-type HRE sequence, in the presence of E. coli purified His-tagged Hoxa2 protein, its Hoxa2(QN-AA) mutant derivative (Hoxa2*) and in vitro translated Pbx1a and Prep1 proteins. Hoxa2 binds to the HRE sequence alone (H), or in combination with Pbx1a (HP). Addition of both Pbx1a and Prep1 generated a similar retarded complex (PP) whatever the wild-type or mutant Hoxa2 protein is involved (see text for comments). The binding assays involving the in vitro translated samples generated aspecific retarded complexes corresponding to the reticulocyte extracts (lysate). (B) Similar experiments were run with oligonucleotides containing a mutated HRE site. While the wild-type sequence (HRE) allowed complex formation with Hoxa2 (H), Hoxa2–Pbx1a (HP) or Pbx1a and Prep1 (PP), no binding was observed on the mutant sequence (HRE*). Assays involving the purified Hoxa2 protein and reticulocyte extracts devoid of expression vectors (TnT) reveal aspecific complex formation (lysate). (C) The HRE sequence is recognized by Hoxa1 and Pbx1a (HP) while not by Hoxa1. The hexapeptide mutant Hoxa1(WM-AA) does not bind the HRE sequence, neither alone nor in combination with Pbx1a. Again, addition of both Pbx1a and Prep1 generates a similarly retarded complex with either the wild-type or mutant Hoxa1 protein (see text for comments). (D) To address the specificity of Hoxa2 binding to the HRE sequence, competition experiments were performed with a 100-fold molar excess of unlabelled wild-type (comp) or mutant oligonucleotides (comp*) with respect to labelled probes. Only the wild-type competitor titrates out both Hoxa2–Pbx1a (HP) and Pbx1a-Prep1 (PP) complex formation. (E) Assays including anti-Hoxa2 or anti-Pbx1 antibodies were performed to confirm the identity of the proteins involved in the shifted complexes. Complex formation with Hoxa2 (H) and Hoxa2–Pbx1a (HP) was impaired by the anti-Hoxa2 antibody, whereas complexes obtained by involving Pbx1a and Prep1 or Pbx1a, Prep1 and Hoxa2 were not. This shows that only dimeric Pbx1a–Prep1 complexes (PP) were formed in the presence of Pbx1a and Prep1 proteins, and that no trimeric complexes including Hoxa2 were obtained. Conversely, the anti-Pbx1 antibody chased and super-shifted the Pbx1a–Prep1 containing complexes (PPIgG).