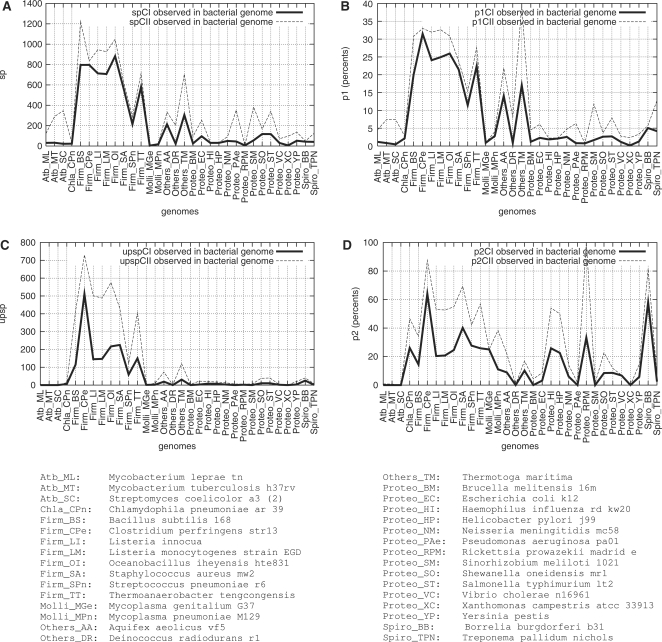
Figure 1.
Frequencies of genes harbouring a putative strong promoter (SP), under four constraint sets, in 32 prokaryotic genomes. See text, Subsection “Genome analysis upon request” for the definition of CI and CII constraints. (A) and (B): UP element optional; (C) and (D): UP element required. Along the x-axis, the following phyla and groups are encountered: Actinobacteria, Chlamydia, Firmicutes (among which Mollicutes), “Others” group, Proteobacteria, Spirochaetales. (A) y-axis: number of genes harbouring a SP (sp); (B) y-axis: ratio p1 of genes harbouring a SP (sp) to the total number of genes encoding proteins in the genome (g), p1 = 100 × sp / g; (C) y-axis: number of genes identified with an UP element harboured in the SP (upsp); (D) y-axis: ratio p2 of the number of genes with an UP element in the SP (upsp) to the number of genes with a SP (sp), p2 = 100 × upsp / sp.