Abstract
N1-methylation of adenosine to m1A occurs in several different positions in tRNAs from various organisms. A methyl group at position N1 prevents Watson–Crick-type base pairing by adenosine and is therefore important for regulation of structure and stability of tRNA molecules. Thus far, only one family of genes encoding enzymes responsible for m1A methylation at position 58 has been identified, while other m1A methyltransferases (MTases) remain elusive. Here, we show that Bacillus subtilis open reading frame yqfN is necessary and sufficient for N1-adenosine methylation at position 22 of bacterial tRNA. Thus, we propose to rename YqfN as TrmK, according to the traditional nomenclature for bacterial tRNA MTases, or TrMet(m1A22) according to the nomenclature from the MODOMICS database of RNA modification enzymes. tRNAs purified from a ΔtrmK strain are a good substrate in vitro for the recombinant TrmK protein, which is sufficient for m1A methylation at position 22 as are tRNAs from Escherichia coli, which natively lacks m1A22. TrmK is conserved in Gram-positive bacteria and present in some Gram-negative bacteria, but its orthologs are apparently absent from archaea and eukaryota. Protein structure prediction indicates that the active site of TrmK does not resemble the active site of the m1A58 MTase TrmI, suggesting that these two enzymatic activities evolved independently.
INTRODUCTION
Transfer RNAs (tRNAs) contain a large number of modified nucleosides generated posttranscriptionally by a variety of enzymes (1). Among nucleoside modifications, base and/or ribose methylations are by far the most frequent and diverse. They are catalyzed by methyltransferases (MTases), a great majority of which use S-adenosyl-l-methionine (AdoMet) as a methyl group donor (2).
The modified nucleoside 1-methyladenosine (m1A) is found at four different positions (9, 14, 22 and 58) in tRNAs (3) and is also formed at position 57 as an intermediate in the biosynthesis of 1-methylinosine (m1I) (4). m1A9 occurs in mammalian mitochondrial tRNAs. The presence of this modification in mitochondrial tRNALys avoids the formation of an alternative structure of the tRNA by preventing the formation of a base pair between A9 and U64 (5). An m1A9 activity was detected in extracts prepared from mitochondrial fractions from human HeLa cells; the corresponding gene, however, has not been identified. m1A14 is rare and thus far has been reported only in cytoplasmic tRNAPhe from several mammals (3). A tRNA:m1A14 MTase activity was identified in an extract prepared from rat brain cortices and the enzyme was partially purified (6). The gene encoding this enzyme also remains unknown. Unlike the two aforementioned positions of m1A modification, m1A58 is found in tRNAs from organisms belonging to the three domains of life. The genes and corresponding enzymes (Trm6 and TrmI in eukaryota and prokaryota, respectively) were identified and characterized in yeast (7) and humans (8), in the bacteria Thermus thermophilus (9) and Mycobacterium tuberculosis (10) and in the archaeon Pyrococcus abyssi (11). The archaeal tRNA:m1A58 MTase was shown to be a region-specific enzyme, also forming m1A at position 57 (intermediate in the formation of m1I). The yeast tRNA:m1A58 MTase is essential for growth, its absence leading to degradation of the initiator tRNA (7). On the other hand, a T. thermophilus mutant, in which the trmI gene has been inactivated, displays a thermosensitive phenotype (9).
In the present work, we focus our attention on the MTase that catalyzes m1A at position 22 in tRNA. So far, this modification has been found only in tRNASer and tRNATyr (tRNAs with a long extra arm) from Bacillus subtilis (12,13) and Mycoplasma capricolum (14). Earlier studies showed that an AdoMet-dependent m1A MTase activity is present in B. subtilis extracts (15). This B. subtilis enzyme modifies adenosine in the sequence 5′-AA22GGC-3′ and requires that the target nucleoside, which is part of the D-loop, is not paired (16). In this work, we demonstrate that this enzyme, hereafter named TrmK, is encoded by the yqfN gene in B. subtilis.
MATERIALS AND METHODS
Bioinformatics analyses
Searches of the non-redundant version of current sequence databases (nr) were carried out at the NCBI using PSI-BLAST (17) with default parameters, except for using a stringent expectation (e)-value threshold of 1e–30 to restrict the analysis to YqfN orthologs and prevent ‘explosion’ of the search by inclusion of all members of the large Rossmann fold methyltransferase (RFM) superfamily. Multiple sequence alignment of the YqfN protein family was calculated using PROMALS (18) with default parameters. Comparisons between the YqfN family and families in the Clusters of Orthologous Groups (COG) database were done with HHpred (19). Based on the YqfN family alignment, a phylogenetic tree was calculated with MEGA 3.1 (20), using the Neighbor-Joining method, with the JTT model of substitutions and pairwise deletions. The stability of individual nodes was calculated using the bootstrap test (1000 replicates).
Secondary structure prediction, identification of ordered and disordered regions and tertiary fold recognition (FR) was carried out via the GeneSilico MetaServer (21), which is a gateway for >30 third-party methods (for references to original methods see http://genesilico.pl/meta2/). Based on the FR alignments to the top-scoring templates we constructed the homology model of YqfN using the so-called ‘FRankenstein's Monster’ method (22, 23), which has been validated as one of the best approaches for template-based modeling in CASP5 and CASP6. The C-terminal extension has been modeled using the ROSETTA method (24) for de novo (template free) protein structure prediction. Model quality has been assessed using PROQ (25). Mapping of sequence conservation onto the model was done via the COLORADO3D server (26), using the Rate4Site method, with the JTT substitution matrix and ML model for rate inference.
Cloning of the B. subtilis yqfN gene
The B. subtilis yqfN gene was amplified by PCR from B. subtilis strain 168 DNA using oligonucleotides yqfn-1/3 and yqfn-2 (Table 1) and Pwo DNA polymerase (Roche Diagnostics, Rotkreuz, Switzerland). The 716 bp amplified product was cloned into the PCRII blunt vector using the PCR cloning kit of (Invitrogen, Carlsbad, USA), giving PCR-yqfN. The cloned fragment was verified by sequencing. The 705 bp NdeI-XhoI fragment of PCR-yqfN was then cloned between the NdeI and XhoI restriction sites of the pET28b expression vector, resulting in the plasmid pET-yqfN. This plasmid allowed T7 expression in Escherichia coli of the B. subtilis YqfN protein bearing an N-terminal His-tag.
Table 1.
Sequence of used oligonucleotides
| Name | Sequence of oligonucleotide | Use |
|---|---|---|
| Yqfn-1 | 5′-TCGACATATGGCCGATATCGGCTCTGAC-3′ | Amplification of a truncated variant of yqfN gene |
| Yqfn-2 | 5′-TCGACTCGAGTTAGCCATGATCGATTACCTCCTTTAACAGCTC-3′ | |
| Yqfn-3 | 5′-TCGACATATGAACGAATTAAAATTATCTAAACGATTG-3′ | Amplification of a full-length variant of yqfN gene |
| Yqfn-2 | 5′-TCGACTCGAGTTAGCCATGATCGATTACCTCCTTTAACAGCTC-3′ | |
| Yqfn-4 | 5′-TACCAAGCTTGGGAGACGGACTGGAAGTG-3′ | Amplification of a central part of yqfN gene |
| Yqfn-5 | 5′-TACGGGATCCGAAAGGGCCAACGAGCATC-3′ | |
| tser-1 | 5′-TATCGCTTAATACGACTCACTATAGGAGAGCTGTCCGAGTGGTCG-3′ | Amplification of tRNASer gene |
| tser-2 | 5′-TATCGCCCTGGTGGCGGAGAGCAAGGGATTCG-3′ | |
| p-ext | 5′ CACGATTTCCAATC-3′ | Primer extension |
Expression and purification of the recombinant YqfN protein
The His-tagged B. subtilis YqfN protein was expressed in the E. coli strain BL21(DE3). This strain transformed by pET-yqfN was grown at 37°C in 2 l of LB supplemented with kanamycin (30 µg/ml) to an optical density of 0.5 at 660 nm. At this stage IPTG was added (1 mM final) to induce recombinant protein expression. Cells were harvested after another 3 h of incubation and resuspended in 20 ml of buffer A (Tris–HCl 50 mM, MgCl2 10 mM, pH 8) and sonicated for 30 min at a power of 200 Watts. The extract was centrifuged for 1 h at 50 000g. The bulk of the nucleic acids were removed by adding 0.15 ml of a 10% polyethyleneimine solution for each milliliter of supernatant of the above step. The solution was stirred on ice for 20 min and centrifuged for 15 min at 15 000g. The pellet was discarded and solid ammonium sulfate was added to 33% saturation. After stirring for 15 min, the precipitate was removed by centrifugation at 18 000g for 15 min. The supernatant was saturated at 90% ammonium sulfate in the same conditions as described above. The pellet was collected by centrifugation at 18 000g for 15 min and dissolved in 8 ml buffer A. The sample was loaded on a Nickel Sepharose chelating column (1.6 × 10 cm) equilibrated in Tris–HCl 50 mM, MgCl2 10 mM and NaCl 500 mM, pH 8 and eluted with a gradient of imidazole from 0 to 1 M in the same buffer. Electrophoretically pure YqfN protein (12 mg) was obtained after chromatography on a Superose P12 molecular sieve column equilibrated with Tris–HCl 50 mM, MgCl2 10 mM, NaCl 100 mM, pH 8.
Cloning and T7 in vitro transcription of the B. subtilis tRNASer (GGA) gene
The sequence encoding the B. subtilis tRNASer (GGA) was amplified by PCR using oligonucleotides tser-1 and tser-2 (Table 1) and Pwo DNA polymerase. The PCR product was cloned into the SmaI site of the pUC18 vector, giving plasmid pUC-tser in which the tRNA sequence is flanked by a 5′ T7 promoter and a 3′ MvaI restriction site. The plasmid insert was verified by sequencing. The general procedure for the generation of in vitro transcripts of tRNA genes is based on the method described previously (27). Radioactive (32P) in vitro transcripts were obtained using MvaI digested pUC-tser plasmid as template. [α-32P]-ATP and [α-32P]-GTP were from ICN Biomedicals, Irvine, USA and T7 RNA polymerase from Roche Diagnostics. Radioactive transcripts were purified by 10% polyacrylamide gel electrophoresis.
tRNA MTase assays
The two types of tRNA MTase assays used in this work were described in (9). The first method consisted in measuring the amount of 14C transferred to total E. coli or B. subtilis yqfN tRNA using [methyl-14C] AdoMet as methyl donor. The reaction mixture (400 μl) consisted of 50 mM Tris–HCl, 10 mM MgCl2, 100 μg total tRNA, 25 nCi [methyl-14C] AdoMet (50 mCi/mmol; GE Healthcare, Piscataway, USA) and extract or enzyme. The second type of tRNA MTase assay involved in vitro transcribed, 32P-labeled tRNAs as substrates (28). Modified nucleotides were analyzed by 2D-TLC on cellulose plates (Merck, Whitehouse Station, USA). First dimension developed with solvent A (isobutyric acid/concentrated NH4OH/water; 66/1/33; v/v/v); second dimension developed with solvent B (0.1 M sodium phosphate pH 6.8/(NH4)2SO4/n-propanol; 100/60/2; v/w/v). The nucleotides were identified using a reference map (29).
yqfN gene inactivation in B. subtilis
B. subtilis yqfN inactivation was performed according to (30), using the pMUTIN integrational vector (conferring erythromycin resistance) in which a 324 bp fragment of the central part of the yqfN gene (yqfNc) was cloned. Integration of this pMUTIN-yqfNc plasmid occurs into the target gene yqfN. The 324 bp yqfNc fragment was PCR amplified using oligonucleotides mut-1 and mut-2 (Table 1) which contain a HindIII and a BamHI restriction site, respectively, to facilitate its cloning into the HindIII and BamHI sites of the pMUTIN vector. The resulting plasmid pMUTIN-yqfNc was transformed into B. subtilis according to (31). Transformants were selected on erythromycin plates (200 mg/l).
Detection of m1A by HPLC
A sample of ∼200 μg of totally hydrolyzed tRNA from the B. subtilis WT and ΔtrmK strain was injected on a Supelco Discovery C18 (250 × 4.6) mm HPLC column equilibrated with ammonium acetate 0.25 M, pH 8. The column was eluted with a linear gradient of acetonitrile/water (40/60) at a flow rate of 1.2 ml/min. The nucleosides were detected by UV absorption at 254 nm.
Mapping of m1A residue
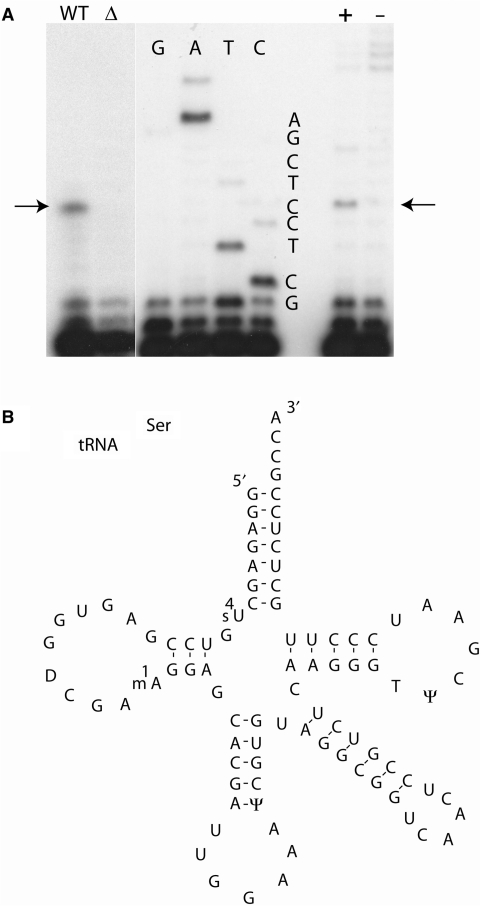
Two different tRNA sources were used. On one hand total tRNA was isolated from WT and ΔtrmK B. subtilis cells. tRNAs with a long extra arm (tRNASer of B. subtilis has an extra arm) were isolated from a 10% polyacrylamide gel. On the other hand B. subtilis tRNASer was synthesized in vitro using the T7 RNA polymerase high yield kit from Fermentas, Vilnius, Lithuania. Two micrograms of these tRNAs were used for hybridization with a 5′ end labeled primer (Table 1). The primer (0.5 pmol) was annealed to the tRNA in 50 mM Tris–HCl, pH 8.6, 60 mM NaCl and 10 mM DTT for 2 min at 90°C and then gradually cooled down to room temperature. Primer extension and sequencing reactions were performed as described in (32).
RESULTS
Prediction that YqfN is the MTase responsible for the formation of m1A22 in tRNA and correction of its amino acid sequence
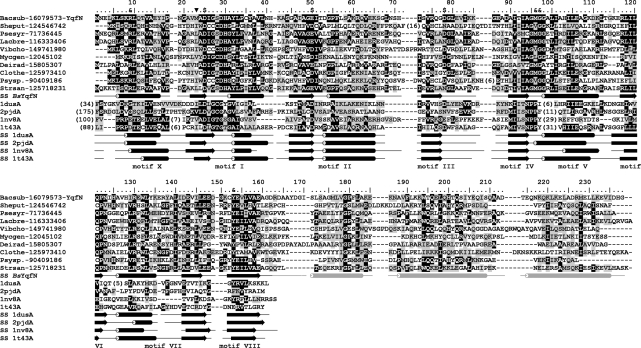
As a part of a large-scale project aiming at identification of new RNA MTases (33) we searched for families from the RFM superfamily (34), whose pattern of phylogenetic occurrence, as indicated in the COG database (35), exhibited similarity to patterns of occurrence of known RNA methylations without known enzymes (1,36). In the course of this analysis, we identified the COG2384 family as potential candidates for the so far uncharacterized enzymes responsible for the biosynthesis of m1A modification present at the position 22 of tRNASer and tRNATyr in B. subtilis and M. capricolum, but not present in E. coli, Mycobacteria and Archaea [among organisms, for which tRNA sequences are available (3)]. Searches of the non-redundant sequence database at the NCBI using PSI-BLAST queried by the consensus sequence derived from COG2384 revealed a family of orthologs conserved in Firmicutes (including Bacillus and Mycoplasma), but absent from E. coli, Mycobacteria and Archaea (Figure 1). Independently, de Crecy-Lagard and coworkers have also found the agreement between the phyletic patterns of m1A22 and COG2384, and proposed that these proteins may be MTases responsible for this modification (37).
Figure 1.
Multiple sequence alignment of the YqfN/TrmK family. Only representative sequences are shown and named using a six-letter abbreviation for genus and species, e.g. Bacsub for B. subtilis, followed by the NCBI GI number. Full alignment is available as Supplementary Data 1. The amino acid residues of B. subtilis YqfN/TrmK are numbered. The template structures detected by FR and used for modeling are shown at the bottom, identified by their PDB accession numbers. Invariant and conserved residues are highlighted in black and dark grey, respectively. Secondary structures taken from the final model of YqfN/TrmK and observed in template structures are indicated as tubes and arrows (helices and strands, respectively). Secondary structure of YqfN/TrmK is colored black in regions based on conserved parts of the templates, while regions folded de novo are shown in light grey. Residues predicted to be involved in AdoMet binding (‘$’) or RNA binding and/or catalysis (‘&’) are indicated above the alignment. The previously proposed (apparently incorrect) N-terminal methionine is indicated by ‘inverted filled triangle’.
As a representative sequence for experimental validation we selected the YqfN protein from B. subtilis because of the relative ease of genetic manipulations with B. subtilis and the availability of tRNA sequences from this organism with experimentally determined position of modified nucleosides (3). However, analysis of the COG2384 multiple sequence alignment (Figure 1) revealed that the YqfN sequence present in the database under Gene Identification (GI) no. 1731002 is N-terminally truncated compared to most of its orthologs. Therefore, we checked for alternative start codons lying upstream from the postulated 5′ end of the yqfN open reading frame (ORF). We found that a tentative translation starting from the alternative codon GTG (66 nucleotides upstream from the one putative ATG codon from the GenBank entry) possesses an N-terminal extension of the YqfN amino acid sequence ‘MNELKLSKRLQTVAEYIPNGAV’ that resembles the typical N-terminus of YqfN orthologs (Figure 1). This finding has been supported by protein structure prediction (see below), which suggested that the N-terminal amino acid sequence of the YqfN that is missing from the variant in GenBank, is involved in the formation of the N-terminal α-helix that might be important both for protein stability and enzymatic activity.
The B. subtilis full-length yqfN gene encodes an MTase involved in the formation of m1A in tRNA (TrmK)
Two versions of the B. subtilis yqfN gene, with or without the 66 nucleotides extension, were PCR amplified and cloned into the pET28 expression vector, allowing the production of N-terminally His-tagged proteins (‘full length’ or ‘truncated’, respectively) in E. coli. After IPTG induction both the full length and the truncated His-tagged proteins showed equal expression in E. coli (data not shown). The MTase activity in extracts of E. coli cells expressing the YqfN proteins was measured at 37°C during 30 min with [methyl-14C] AdoMet and unfractionated E. coli tRNA as a substrate. Crude E. coli tRNA was used, since it does not contain m1A22. Moreover, it was previously shown that an MTase present in B. subtilis extract could catalyze the formation of m1A in the D-loop of E. coli tRNA (16). Incorporation of 14C-methyl into E. coli tRNA was only detected in the presence of the extract containing the full-length enzyme. The 22 N-terminal amino acids missing from the original YqfN sequence in the database are thus important for the MTase activity of YqfN. We propose to rename YqfN as TrmK, according to the traditional nomenclature for bacterial tRNA MTases, or TrMet(m1A22) according to the nomenclature from the MODOMICS database of RNA modification enzymes.
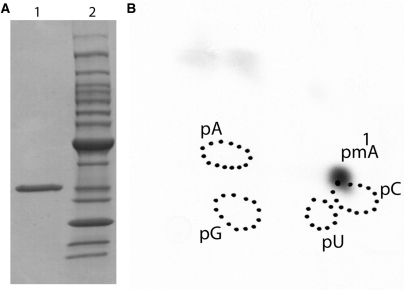
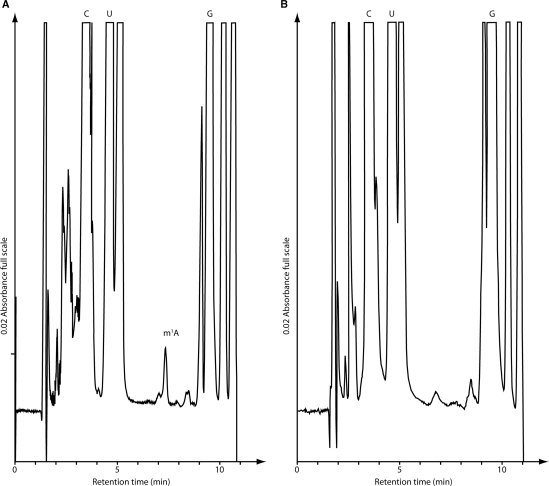
The His-tagged full-length TrmK protein was purified to homogeneity by ammonium sulfate precipitation followed by immobilized metal ion affinity chromatography (Figure 2A). The activity of the purified enzyme was measured using different kind of substrates. First, unfractionated E. coli tRNA was used. After incubation of the purified enzyme with E. coli tRNA and [methyl-14C] AdoMet, the tRNA was recovered by phenol extraction and ethanol precipitation and completely hydrolyzed into 5′-phosphate nucleosides by nuclease P1. The resulting hydrolyzate was analyzed by 2D-TLC followed by autoradiography. The results shown in Figure 2B revealed the presence of a single radioactive compound with migration characteristics identical to that of 1-methyladenosine 5′-phosphate (pm1A) according to the published reference map (29). Moreover, tRNA isolated from the B. subtilis strain in which the trmK gene was inactivated (see Materials and Methods section) appeared to be a good substrate for the enzyme (which indicates that this tRNA lacks m1A22), in contrast to tRNA isolated from WT B. subtilis cells (which has m1A22 and hence cannot be further modified). Indeed, HPLC analyses showed that m1A was absent in tRNA isolated from ΔtrmK B. subtilis cells (Figure 3).
Figure 2.
Affinity-purified B. subtilis TrmK protein catalyzes the formation of m1A in E. coli tRNA in vitro. (A) SDS–PAGE (Biorad, Hercules, USA; 4–20% acrylamide) analysis under reducing conditions of purified B. subtilis TrmK protein. Lane 1, 6.5 μg of purified protein. Lane 2, molecular bench marker standard from Invitrogen. (B) Autoradiography of a 2D-chromatogram of 5′-phosphate nucleosides on thin layer cellulose plate. Total E. coli tRNA was incubated in the presence of [methyl-14C] AdoMet and 5 μg of purified YqfN protein as described in Materials and Methods section. After incubation at 37°C the tRNA was recovered, digested by nuclease P1 and the resulting nucleotides were analyzed by 2D-TLC on a cellulose plate. Circles in dotted lines show the migration of the four canonical nucleotides as UV markers.
Figure 3.
Detection of nucleosides in B. subtilis tRNA. (A) tRNA from B. subtilis WT strain and (B) tRNA from ΔtrmK B. subtilis strain. The column was calibrated with authentic samples of C, cytidine; U, uridine; G, guanosine and m1A, 1-methyladenosine.
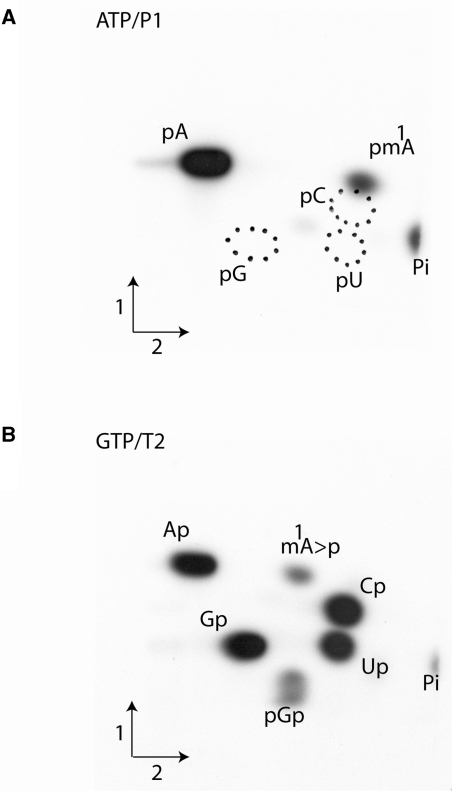
Further evidence for the formation of m1A catalyzed by the TrmK protein was obtained using a second type of experiment. The purified enzyme was incubated under identical experimental conditions as above but with either an [α-32P]ATP or an [α-32P]GTP labeled precursor tRNA substrate obtained after in vitro transcription by T7-RNA polymerase of a synthetic B. subtilis tRNASer(GGA) gene. After the incubation with purified TrmK and AdoMet, the formation of m1A in the B. subtilis tRNASer was analyzed by 2D-TLC. The [α-32P]ATP-labeled tRNASer was completely hydrolyzed with nuclease P1 to generate 5′-phosphate nucleosides with the [32P]-phosphate present only in 5′-phosphate adenosine and 5′-phosphate adenosine derivatives (Figure 4A), while the [α-32P]-GTP labeled tRNA was digested with RNAse T2, thus generating the different 3′-phosphate nucleosides of which only those that were 5′-adjacent to G in the tRNA sequence harbored a 32P-radiolabeled phosphate (nearest neighbor analysis; Figure 4B). The results show the presence of m1A, 5′-adjacent to a G in the B. subtilis sequence. Note that hydrolysis by RNAse T2 resulted in the accumulation of 1-methyladenosine 2′-3′-cyclic phosphate (m1A > p; an intermediate in the hydrolysis reaction) because the presence of this modification inhibits the action of RNAse T2, as has already been reported (9). Quantification of the relative amount of [32P] in the different radioactive spots on the TLC plates, revealed that ∼1 mol of m1A is formed per mole of tRNA after 30 min of reaction in the presence of 5 μg of enzyme.
Figure 4.
Characterization of the tRNA MTase activity of recombinant TrmK protein using radiolabeled in vitro transcripts of B. subtilis tRNASer. The transcripts were incubated in the presence of 5 μg of B. subtilis TrmK enzyme and AdoMet. The transcripts were then digested by either nuclease P1 (A) or RNase T2 (B) and the resulting nucleotides were analyzed by 2D-TLC on cellulose plates and autoradiography. The nature of the labeled triphosphate nucleoside and the enzyme used to hydrolyze the transcripts are given above the autoradiographs. Circles in dotted lines show the migration of the canonical nucleotides used as UV markers. m1A > p is for 1-methyladenosine 2′–3′ cyclic phosphate.
The TrmK protein catalyzes the formation of m1A at position 22 of tRNA
The nearest neighbor analysis above shows that the m1A formed by TrmK is 5′ adjacent to a G in B. subtilis tRNASer. Since in this tRNA there are several 5′-AG-3′ pairs, primer extension experiments were performed to determine the precise position in the tRNA of the m1A modification. This was determined for tRNA modified in vivo and in vitro. In the in vitro experiment, a tRNASer transcript was incubated in the presence of AdoMet and the TrmK enzyme for 30 min at 37°C. This modified transcript as well as the unmodified transcript was used for a primer extension experiment, using an oligonucleotide complementary to nucleotides 29–42 of B. subtilis tRNASer. Figure 5A (lane +) shows a transcription stop at position 23 of the tRNASer, consistent with the presence of m1A at position 22. In the in vivo experiment, tRNA was isolated from B. subtilis WT strain and from ΔtrmK B. subtilis strain (for the construction of this latter see Materials and Methods section) and were used in primer extension experiments using the same oligonucleotide as above. The results presented in Figure 5A (lane WT) show a strong stop at position 23. Thus, m1A is present at position 22 of tRNASer of the B. subtilis WT strain.
Figure 5.
Localization of m1A in B. subtilis tRNASer. Primer extension was performed with a primer complementary to nucleotides 29–42 of tRNASer of B. subtilis, synthesized in vitro or in vivo. (A) The arrow indicates a strong stop at m1A. The in vitro synthesized T7 transcript of tRNASer was incubated with (+) or without (−) the YqfN enzyme. B. subtilis tRNA was isolated from WT or ΔtrmK B. subtilis cells. (B) Shows the sequence of the tRNASer (GGA) of B. subtilis.
Growth is not impaired in a ΔtrmK B. subtilis strain
To check whether there is a phenotype linked to the absence of m1A22 in B. subtilis tRNA, different culture conditions were analyzed. Growth at 37, 45 or 55°C was not affected by the mutation. In a competition experiment where both type of cells were initially inoculated in equal amounts, the cell distribution remained unchanged for 70 generations (measured by replica plating on plates with and without erythromycin). Thus, growth was not impaired by the trmK mutation. Earlier studies showed a relationship between tRNA modification and sporulation in B. subtilis (38,39). Therefore, we have compared sporulation frequency between the WT and ΔtrmK B. subtilis cells using the methodology described in (38). No difference in the sporulation efficiency was observed (data not shown).
Phylogenetic analysis of the TrmK family
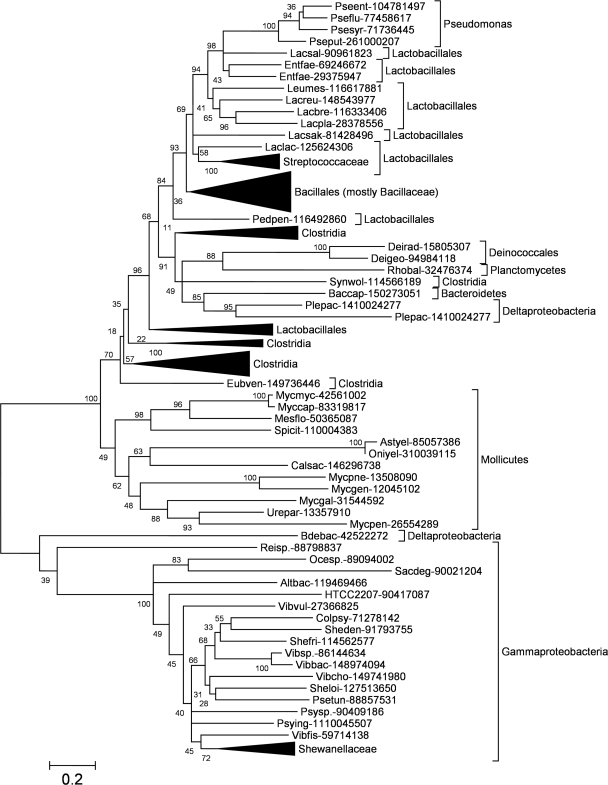
Despite the absence of COG2384 family members in those Gammaproteobacteria, for which no m1A was detected in tRNA, they are present in some Gammaproteobacteria, in particular in Alteromonadales, Vibrio and Pseudomonas. Thus, a question arises about the details of phylogenetic distribution and evolutionary history of these proteins. A phylogenetic tree of the COG2384 family (Figure 6) reveals that these proteins split into two main lineages, one grouping together mostly Gram-positive, and the other exclusively Gram-negative bacteria (Gammaproteobacteria and just a single Deltaproteobacterium). The ‘Gram-positive’ lineage can be further subdivided into two branches. One branch comprises only Mollicutes, including M. capricolum, for which there is experimental evidence of m1A22 methylation in tRNA by the so far uncharacterized enzyme (14). The other branch includes Clostridia (divided into several groups, scattered at the base of the branch with no bootstrap support for their unification into a monophyletic group), Lactobacillaceae (also split into several branches) and Bacillales (apparently monophyletic grouping, but with very weak bootstrap support). The ‘Gram-positive’ lineage includes two groups of Gram-negative members from Pseudomonas and Deltaproteobacteria. According to the bootstrap support of the calculated topology COG2384 genes have been acquired by their Gram-negative hosts due to horizontal gene transfers: independently from Lactobacillales to Pseudomonas and from Clostridia or Lactobacillales to Deltaproteobacteria. Thus, our phylogenetic analysis suggests that either COG2384 is an ancient bacterial family, whose members have been lost from most Gram-negative species such as E. coli (and have been reintroduced later to some species by horizontal gene transfer) or that it originated in Firmicutes (in which it is now most prevalent) and has been transferred horizontally to other species.
Figure 6.
Phylogenetic tree of the TrmK family. Sequences are indicated by the abbreviated genus and species name (e.g. Bacsub for B. subtilis) and the NCBI GI number and their phylogenetic origin are indicated. For clarity of the presentation, branches with sequences belonging to the same taxa have been collapsed and are shown as triangles. Values at the nodes indicate the statistical support for the particular branches, according to the bootstrap test.
Comparison of the TrmK family with other MTase families using a rigorous method for the detection of remote homology (HHpred) revealed no close relationship between TrmK and any other MTase family, and only comparable similarity to various RFM fold MTases with different specificities, e.g. PrmA, CobL, RsmC and PrmC (Supplementary Table 1). Closer inspection of family–family alignments revealed potential conservation of the common cofactor-binding site (sequence motifs I–III) between TrmK and the above-mentioned MTases, however, indicated significant differences between their putative catalytic centers (sequence motifs IV, VI and VIII), consistent with different substrate specificities of these enzymes. In the absence of similarity of active sites at the sequence level, we turned to protein structure prediction in order to identify a likely active site of TrmK from the analysis of conserved residues that may form a binding pocket in three dimensions.
Structure prediction of the B. subtilis TrmK protein
Bioinformatics analysis of the B. subtilis TrmK sequence revealed that residues 1–162 show a propensity to form a well-folded globular domain rich in α-helices and β-strands, while the C-terminal residues 163–238 exhibit a tendency to form a coiled-coil structure with three predicted α-helices. We constructed a 3D model of TrmK using the top-scoring structures 1dus and 1ufk as templates for residues 1–162, while the C-terminal extension has been folded de novo (see Materials and Methods section for details). The full-length model of TrmK (shown in Figure 7) has been evaluated by PROQ as potentially ‘very good’ (predicted LGscore of 3.777). Finally, the methyl group donor AdoMet has been docked into the predicted active site in the globular domain by analogy to the known structures of RFM proteins.
Figure 7.
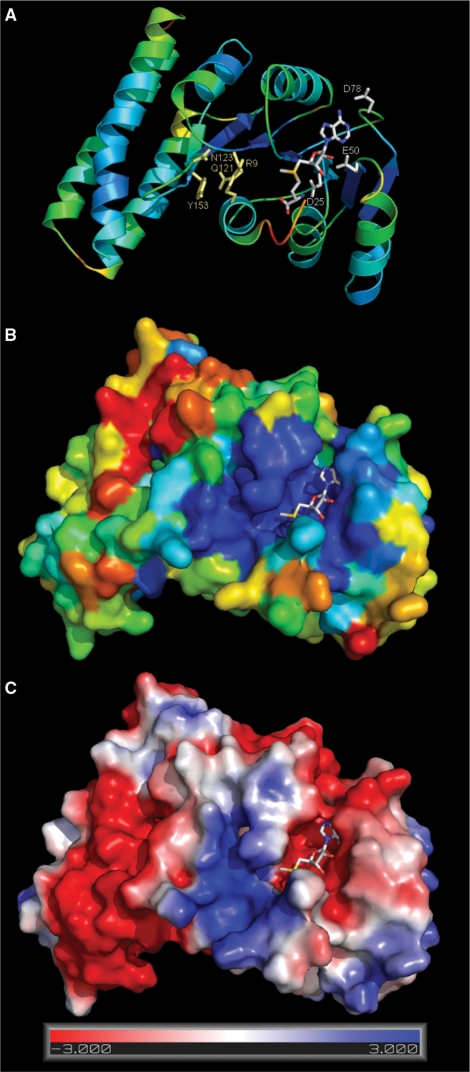
Structural model of B. subtilis TrmK. Coordinates of the model are available from ftp://genesilico.pl/iamb/models/TrmK/ (A) TrmK model colored according to the predicted accuracy (estimated agreement with the native structure), from blue (highly confident, predicted error ∼1 Å), to yellow (predicted low accuracy, expected differences between the model and the native structure up to ∼5 Å), to red (predicted low accuracy, error >5 Å). (B) TrmK model in the surface representation, colored according to sequence conservation in the TrmK family, from deep blue (invariant), to light blue (conserved), to yellow/red (highly variable). A highly conserved blue patch indicates the cofactor-binding site and the predicted tRNA-binding/catalytic site. (C) TrmK model colored according to the distribution of electrostatic potential, from red (−3 kT) to blue (+3 kT). AdoMet binds in a negatively charged, red-colored cleft at the right-hand side, while a blue patch in the middle of the structure suggests the localization of a tRNA-binding site around the catalytic pocket.
Analysis of the electrostatic potential distribution reveals that the TrmK protein surface around the putative cofactor-binding pocket is negatively charged, compatibly with the positive charge of the cofactor, while the putative substrate-binding site is positively charged, compatibly with the negative charge of the RNA phosphate backbone. Mapping of the sequence conservation onto the protein structure reveals concentration of conserved residues in the predicted catalytic pocket of the N-terminal domain, and high variability in the helical C-terminus. By analogy to other members of the RFM superfamily, and based on the analysis of sequence conservation among TrmK homologs, we predict that residues D25, E50 and D78 are involved in coordination of the methionine, ribose and adenine moieties of AdoMet, respectively. A nearly invariant pair of residues in motif VII, corresponding to R130 and E143 in the B. subtilis TrmK sequence, may form a salt bridge potentially important for protein folding and/or stability.
Based on the model we predict that the side chains of conserved residues R9 (in motif X), and Y153 (motif VIII) are involved in substrate binding and/or catalysis. Conserved residues G95 and G97 appear to be important for positioning of a hydrophobic residue at position 96 (Met in B. subtilis TrmK), which may be important for stabilizing the target base by van der Waals interactions. A conserved residue Q121 (motif VI) may be involved in binding of the target base. However, it is substituted by Glu in TrmK homologs from some of the Lactobacilli or by Cys in some members from Mycoplasma and most of those from the Proteobacterial branch. This suggests that Q121 it is unlikely to be directly involved in catalysis, unless the above mentioned TrmK family members exhibit differences in their catalytic mechanism. Interestingly, Q121 of B. subtilis TrmK is homologous to the catalytic Cys in e.g. tRNA:m5C MTases (40). Summarizing, comparison of the predicted TrmK active site with the active site of other MTases, including m1A MTase TrmI acting on a different position 58 in tRNA (9,11), reveals no obvious similarities.
DISCUSSION
The modified nucleosides in tRNA function usually by performing structural tasks, e.g. by blocking or reinforcing the ability of the base to form particular pairs with other bases (41). Addition of a single methyl group at one of the Watson–Crick positions, e.g. N1 atom of adenosine, can bring about a large conformational rearrangement, by preventing formation of a single base pair, as demonstrated for m1A9 modification in human mitochondrial tRNALys (42). A physicochemical understanding of modified nucleoside contributions to the tRNA structure and function is greatly facilitated by the knowledge of these enzymes, which allows easy generation of tRNAs lacking individual modifications by simple genetic techniques. For the model organisms E. coli and Saccharomyces cerevisiae nearly all tRNA modification enzymes have been identified to date (1). However, this is not the case for other organisms, which contain modifications at different positions. One example of a modification enzyme that has remained unidentified is the one responsible for formation of m1A at position 22 of tRNA. In this work, we demonstrated that this enzyme, hereafter named TrmK belongs to the COG2384 protein family, and is encoded by the yqfN ORF in B. subtilis. We found that the published sequence of YqfN contains a truncation of the N-terminus, and we cloned both the original (‘truncated’) and extended (‘full length’) versions and validated the extended variant (but not the original one) for the ability to introduce m1A methylation at position 22 in tRNA.
The trmK mutant shows no detectable phenotype, neither at the growth nor at the sporulation levels. This is however not exceptional since recent work by the group of Phizicky showed that the cumulation of mutations affecting nucleoside modification in tRNA are often necessary to confer a phenotype (43,44). Thus, analyses of other tRNA modification enzymes from B. subtilis and construction of respective mutants are required to elucidate the role of m1A22 modification.
We predict that the catalytic domain of TrmK exhibits a novel type of the active site, without obvious similarities to any known RNA MTase family, including the previously characterized tRNA:m1A58 MTase TrmI. While both enzymes bind the methyl group donor AdoMet in a very similar way, the methyl group acceptor adenosine from tRNA is probably recognized using a completely different mechanism. It is intriguing that the putative active site of TrmK that must bind the positively charged adenine contains a conserved positively charged Arg9, but no negatively charged residue that could facilitate the transfer of a methyl group from a positively charged AdoMet molecule. Unfortunately, the limited accuracy of the current model makes the small molecule docking analysis unfeasible, hence speculations about the catalytic mechanism of m1A22 methylation must await biochemical analyses of the residues highlighted by the model—in particular R9, Q121, Y153 and M96.
Phylogenetic analysis reveals that TrmK has orthologs not only in Gram-positive, but also in Gram-negative bacteria. Interestingly, some of the Gram-negative orthologs of TrmK (e.g. Shewanella and Vibrio) exhibit different amino acid residues in the putative active site, suggesting possible variation in substrate specificity. Therefore, we hope our discovery of the gene encoding the m1A22 MTase and characterization of the respective enzyme in B. subtilis will stimulate comparative experimental analysis of its homologs from other species. We would also like to underscore the value of experimental determination of tRNA sequences (including mapping of modified residues that obviously cannot be made at the level of tDNA sequencing), and would like to encourage scientists from the tRNA field to systematically determine tRNA sequences for organisms representative for different branches of the Tree of Life.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Stéphane Aymerich for the gift of the pMUTIN plasmid and for advice for B. subtilis gene inactivation, and Valérie de Crécy-Lagard and Henri Grosjean for helpful discussions. Bart Scherens is acknowledged for help in the artwork. J.M.B. and K.H.K. were supported by a grant N301 2396 33 and K.L.T. was supported by a Ph.D. grant N301 105 32/3599 from the Polish Ministry of Science. L.D. was supported by grants from the FRFC (Fonds pour la Recherche Fondamentale Collective) and IISN (Institut Interuniversitaire des Sciences Nucléaires), by the Fonds E. Defay, the Fonds Jean Brachet Recherche and the Fonds D. et A. Van Buuren (ULB). Funding to pay the Open Access publication charges for this article was provided by the FRFC (Fonds pour la Recherche Fondamentale Collective).
Conflict of interest statement. None declared.
REFERENCES
- 1.Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozbial PZ, Mushegian AR. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005;5:19. doi: 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosjean H, Auxilien S, Constantinesco F, Simon C, Corda Y, Becker HF, Foiret D, Morin A, Jin YX, Fournier M, et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- 5.Helm M, Attardi G. Nuclear control of cloverleaf structure of human mitochondrial tRNA(Lys) J. Mol. Biol. 2004;337:545–560. doi: 10.1016/j.jmb.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Salas CE, Dirheimer G. In vitro methylation of yeast tRNAAsp by rat brain cortical tRNA-(adenine-1) methyltransferase. Nucleic Acids Res. 1979;6:1123–1133. doi: 10.1093/nar/6.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varshney U, Ramesh V, Madabushi A, Gaur R, Subramanya HS, RajBhandary UL. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res. 2004;32:1018–1027. doi: 10.1093/nar/gkh207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menichi B, Arnold HH, Heyman T, Dirheimer G, Keith G. Primary structure of Bacillus subtilis tRNAsTyr. Biochem Biophys. Res. Commun. 1980;95:461–467. doi: 10.1016/0006-291x(80)90760-3. [DOI] [PubMed] [Google Scholar]
- 13.Matsugi J, Jia HT, Murao K, Ishikura H. Nucleotide sequences of serine tRNAs from Bacillus subtilis. Biochim. Biophys. Acta. 1992;1130:333–335. doi: 10.1016/0167-4781(92)90448-9. [DOI] [PubMed] [Google Scholar]
- 14.Andachi Y, Yamao F, Muto A, Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 1989;209:37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- 15.Raettig R, Kersten H, Weissenbach J, Dirheimer G. Methylation of an adenosine in the D-loop of specific transfer RNAs from yeast by a procaryotic tRNA (adenine-1) methyltransferase. Nucleic Acids Res. 1977;4:1769–1782. doi: 10.1093/nar/4.6.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersten H, Raettig R, Weissenbach J, Dirheimer G. Recognition of individual procaryotic and eucaryotic transfer-ribonucleic acids by B. subtilis adenine-1-methyltransferase specific for the dihydrouridine loop. Nucleic Acids Res. 1978;5:3033–3042. doi: 10.1093/nar/5.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 19.Soding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 21.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosinski J, Cymerman IA, Feder M, Kurowski MA, Sasin JM, Bujnicki JM. A “FRankenstein's monster” approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins. 2003;53(Suppl. 6):369–379. doi: 10.1002/prot.10545. [DOI] [PubMed] [Google Scholar]
- 23.Kosinski J, Gajda MJ, Cymerman IA, Kurowski MA, Pawlowski M, Boniecki M, Obarska A, Papaj G, Sroczynska-Obuchowicz P, Tkaczuk KL, et al. FRankenstein becomes a cyborg: the automatic recombination and realignment of fold recognition models in CASP6. Proteins. 2005;61(Suppl. 7):106–113. doi: 10.1002/prot.20726. [DOI] [PubMed] [Google Scholar]
- 24.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 25.Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasin JM, Bujnicki JM. COLORADO3D, a web server for the visual analysis of protein structures. Nucleic Acids Res. 2004;32:W586–W589. doi: 10.1093/nar/gkh440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes VM, Abelson J. A synthetic substrate for tRNA splicing. Anal. Biochem. 1987;166:90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- 28.Droogmans L, Grosjean H. 2′-O-methylation and inosine formation in the wobble position of anticodon-substituted tRNA-Phe in a homologous yeast in vitro system. Biochimie. 1991;73:1021–1025. doi: 10.1016/0300-9084(91)90143-o. [DOI] [PubMed] [Google Scholar]
- 29.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 30.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144 (Pt 11):3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 31.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006;34:4293–4301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bujnicki JM, Droogmans L, Grosjean H, Purushothaman SK, Lapeyre B. Bioinformatics-guided identification and experimental characterization of novel RNA methyltransferases. In: Bujnicki JM, editor. Practical Bioinformatics. Vol. 15. Berlin: Springer-Verlag; 2004. pp. 139–168. [Google Scholar]
- 34.Bujnicki JM. Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol. 1999;1:175–182. [PubMed] [Google Scholar]
- 35.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Crecy-Lagard V. In: Practical Bioinformatics. Bujnicki JM, editor. Vol. 15. Berlin: Springer-Verlag; 2004. pp. 169–190. [Google Scholar]
- 37.de Crecy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 38.Menichi B, Heyman T. Study of tyrosine transfer ribonucleic acid modification in relation to sporulation in Bacillus subtilis. J. Bacteriol. 1976;127:268–280. doi: 10.1128/jb.127.1.268-280.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buu A, Menichi B, Heyman T. Thiomethylation of tyrosine transfer ribonucleic acid is associated with initiation of sporulation in Bacillus subtilis: effect of phosphate concentration. J. Bacteriol. 1981;146:819–822. doi: 10.1128/jb.146.2.819-822.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helm M, Giege R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- 43.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 44.Kotelawala L, Grayhack EJ, Phizicky EM. Identification of yeast tRNA Um(44) 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNA(Ser) species. RNA. 2008;14:158–169. doi: 10.1261/rna.811008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.