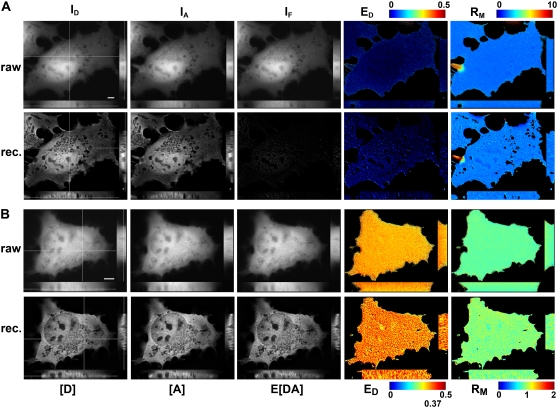
FIGURE 5.
3DFSR provides improved 3D resolution while accurately estimating ED, EA, and RM for uniformly distributed bound and free molecules. The concentrations of [D], [A], and E[DA] were reconstructed by 3DFSR from widefield images of live cells expressing uniform free CFP and YFP (A) or a linked CFP-YFP molecule (B). 3D stacks of ID, IA, and IF from cells expressing free CFP and YFP were used for direct calculation of FRET stoichiometry (top row, ED and RM, EA images were very similar to ED). Thirty iterations of 3DFSR (including photobleaching correction) produced improved estimates of [D] and [A], whereas the reconstructed estimates for E[DA] and ED indicated no complex. Furthermore, RM was precisely maintained. Cells expressing a linked construct demonstrated a uniform ED of ∼0.37 and RM ∼1.0. 3DFSR (including photobleaching correction) generated improved estimates for [D], [A], and E[DA] and accurately returned ratio images with values of 0.37 and 1.0 in all planes. The effects of FRET-enhanced photobleaching and its correction can be observed by comparing the raw ED and reconstructed ED in (B). Scale bar is 5 μm.