Abstract
Selective serotonin reuptake inhibitors (SSRIs) can safely and successfully treat major depression, although a substantial number of patients benefit only partially or not at all from treatment. Genetic polymorphisms may play a major role in determining the response to SSRI treatment. Nonetheless, it is likely that efficacy is determined by multiple genes, with individual genetic polymorphisms having a limited effect size. Initial studies have identified the promoter polymorphism in the gene coding for the serotonin reuptake transporter as moderating efficacy for several SSRIs. The goal of this review is to suggest additional plausible polymorphisms that may be involved in antidepressant efficacy. These include genes affecting intracellular transductional cascades; neuronal growth factors; stress-related hormones, such as corticotropin-releasing hormone and glucocorticoid receptors; ion channels and synaptic efficacy; and adaptations of monoaminergic pathways. Association analyses to examine these candidate genes may facilitate identification of patients for targeted alternative therapies. Determining which genes are involved may also assist in identifying future, novel treatments.
Keywords: pharmacogenetic, pharmacogenomic, depression, SSRI
Introduction
Major depressive disorder (MDD) is a complex disorder associated with various monoaminergic disturbances, abnormalities of endocrine regulation, alterations in sleep physiology, neuroanatomic abnormalities, and neurophysiologic changes (Drevets 1998; Thase 1998; Manji et al 2001; Ordway et al 2002; Moretti et al 2003; Sheline 2003; Buysse 2004). Selective serotonin reuptake inhibitors (SSRIs) are frequently used to treat MDD; however, it is now clear that a large percentage of patients suffering from MDD may not benefit from SSRI treatment (Thase 2003). Evidence from family studies indicates that response to specific antidepressant treatments can be affected by genetic differences (Pare and Mack 1971; O’Reilly et al 1994; Serretti et al 1998). Pharmacogenetics holds the potential to genetically predict who will and will not benefit from SSRIs (Catalano 1999; Pickar and Rubinow 2001; Serretti et al 2002). Moreover, delineating the genes that are associated with poor response to an SSRI may be valuable in identifying alternative patient-specific treatments, such as novel treatments or currently known and available therapies.
Until recently, the focus of pharmacogenetics has been on functional gene variants that may contribute to variable antidepressant exposure (ie, pharmacokinetics). Extensive information is now available regarding genetic variation in many important metabolic enzymes (for reviews see Brosen 1993; Cohen and De Vane 1996; Steimer et al 2001; Daly 2003; Evans and McLeod 2003; Mancama and Kerwin 2003). This review will primarily focus on the putative central mechanisms of SSRI activity and what is known about genetic variants that moderate those pathways. It will also discuss the selection of candidate genes based on these pathways. An extensive critique of individual studies will not be provided, but attention will be given to highlighting the range of potential candidate genes that can be culled using this strategy.
Linkage analysis in humans to identify the genetic loci that affect treatment response has been impractical. Consequently, association analyses are essential (Tabor et al 2002). Linkage analysis does not require preselection of candidate genes, but it does require examining several sets of patients and family members who are treated similarly for depression and who are assessed using similar outcome measures (Kopnisky et al 2002; van den Bree and Owen 2003): something that is exceedingly rare.
Population-based association analyses test for associations of causal candidate polymorphisms or candidate polymorphisms that are tightly linked to a casual polymorphism. Subjects do not need to be genetically related. However, given that there may be over 30 000 genes and several million polymorphisms in humans (Sachidanandam et al 2001; Venter et al 2001; Botstein and Risch 2003), densely screening the genome may require testing up to a million polymorphisms per individual (Botstein and Risch 2003). Thus, rather than screen the genome, it is currently more feasible to select a set of specific candidate genes for study. Many of these genes may interact in complex ways to affect treatment response: synergistically, antagonistically, and additively (Murphy et al 2003). Therefore, a targeted and focused approach for selecting multiple candidate genes for association analyses is essential.
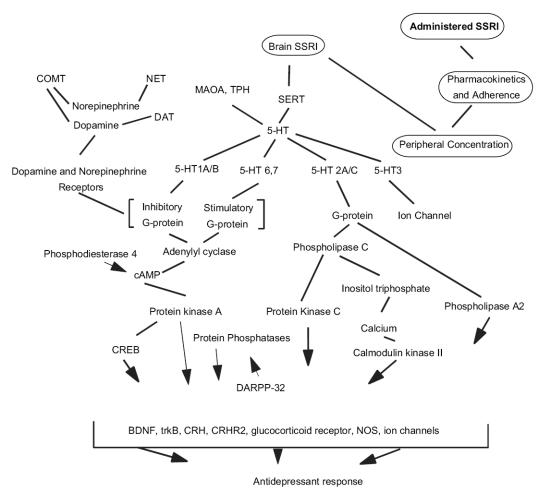
Several strategies exist for selecting candidate genes for study (see Table 1). One is to focus on mechanisms of action. For example, it is now well established that SSRIs inhibit the serotonin transporter (SERT), acutely increasing serotonin (5-HT) within the synapse. 5-HT then binds to several receptors, triggering subsequent intracellular signaling events through G-proteins (Shelton 2000). Genetic differences influencing any of these initial and intracellular signaling events may alter the efficacy of SSRI treatment (Figure 1). A related strategy focuses on putative downstream effects of SSRIs, such as alterations in growth factors and synaptic remodeling, and the effects on synaptic transmission, other transmitter systems, and/or glucocorticoids and corticotropin-releasing hormone (CRH).
Table 1.
Strategies for selecting candidate genes for antidepressant response
| Enzymes and transporters affecting pharmacokinetics and disposition |
| Potential mechanism of antidepressant action (proximally and downstream) |
| Candidate genes for major depression and mood disorders |
| Candidate loci from linkage studies in animal models |
| Expression studies (mRNA and protein) to identify antidepressant-related genes |
| Confirm preliminarily identified candidate genes |
Figure 1.
Potential cascade of events mediating and moderating SSRI effects. The multiple inter-regulatory and feedback pathways are absent from this figure.
Abbreviations: BDNF, brain-derived neurotrophic factor; COMT, catecholamine-O-methyl-transferase; CRH, corticotropin-releasing hormone; CRH2, CRH receptor2; cAMP, cyclic adenosine monophosphate; CREB, cAMP response element binding protein; DARPP-32, dopamine and cAMP regulated phosphoprotein; DAT, dopamine transporter; MAOA, monoamine oxidase A; NOS, nitric oxide synthase; NET, norepinephrine transporter; 5-HT, serotonin; SERT, serotonin transporter; SSRI, selective serotonin reuptake inhibitor; TPH, tryptophan hydroxylase
Another strategy is to identify genes potentially involved in the pathophysiology of major depression and related mood disorders such as bipolar depression (Table 2). Although the genes affecting treatment response are not necessarily the same as those increasing vulnerability to various types of depression, they are biologically plausible targets for investigation.
Table 2.
Candidate loci with preliminary evidence for involvement in affective disorders based on association analyses
| Gene | General function | Candidate polymorphisms | Chromosome allocation |
|---|---|---|---|
| SLC6A4 | 5-HT reuptake | Promoter variable repeata | 17q11.1-q12 |
| TPH1 | 5-HT synthesis | A218Cb | 11p15.3-p14 |
| MAOA | 5-HT catabolism | Promoter variable repeatb | Xp11.4-911.3 |
| MTHFR | Monoamine synthesis | C677T | 1p36.3 |
| HTR1A | 5-HT receptor | C(–1019)G | 5q11.2-q13 |
| HTR1B | 5-HT receptor | C861G | 6q13 |
| A(–161)T | |||
| HTR2A | 5-HT receptor | G(–1438)A | 13q14-q21 |
| C102Tb | |||
| HTR2C | 5-HT receptor | C(–759)T | Xq24 |
| HTR3A | 5-HT receptor | C178T | 11q23.1 |
| HTR6 | 5-HT receptor | C267T | 1p36-p35 |
| GNAS1 | G-protein subunit – alpha | T131C | 20q13.2-q13.3 |
| GNB3 | G-protein subunit – beta | C825Tb | 12p13 |
| PLA2 | Phospholipase A2 | Trinucleotide repeat | 1p36.13 |
| BDNF | Neuronal growth factor | A758G | 11p13 |
| NOS1 | Neuronal nitric oxide synthesis | C29Tb | 12q24.2-q24.31 |
| CRH | Corticotropin-releasing hormone | T1273C | 8q13 |
| CRHR2 | CRH receptor | G1047A | 7p15.1 |
| SLC6A3 | Dopamine transporter | Promoter variable repeat | 5p15.3 |
| SLC6A2 | Norepinephrine transporter | Promoter variable repeat | 16q12.2 |
| ADRB1 | Norepinephrine beta 1 receptor | G145A | 10q24-q26 |
| G1165C | |||
| ADRA2A | Norepinephrine alpha 2 receptor | C(–1291)G | 10q24.q26 |
| COMT | Monoamine metabolism | G158A | 22q11.21-q11.23 |
| DRD2 | Dopamine 2 receptor | –141C insertion/deletion | 11q23 |
| CHRM2 | Muscarinic 2 receptor | A1890T | 7q31-q35 |
| APOE | Apolipoprotein E | C3937Tb | 19q13.2 |
| C4075Tb |
Multiple studies replicate an association with SSRI treatment response.
Polymorphism has a potential preliminary association with antidepressant treatment response in previous studies.
See text for citations. Gene nomenclature is according to HGNC (www.gene.ucl.ac.uk/nomenclature).
Genetic studies using rodent models of depression are capable of identifying novel candidate genes. For example, the linkage of antidepressant response to various chromosomal regions can be examined in selectively bred and recombinant inbred lines of mice. Candidate genes may be selected from these chromosomal regions; a strategy that is being employed for genes affecting sensitivity to drugs of abuse (Buck 1995; Phillips 1997; Fehr et al 2002). Similarly, pharmacogenomic and pharmacoproteomic studies in which the expression levels of genes (either measuring mRNA or the protein itself) are evaluated following SSRI treatment, may be useful in identifying novel genes that are important in mediating SSRI effects (Yamada et al 2000; Landgrebe et al 2002).
This review will highlight candidate genes that are selected based on known or prominently postulated mechanisms of SSRI action, including the initial effects on synaptic serotonin, intracellular events, and the downstream effects on growth factors and other neurotransmitter systems (Figure 1). Supplementary evidence for the involvement of genes or polymorphisms (Table 2) in depression and related mood disorders will also be noted.
5-HT transporter and 5-HT availability
Several proteins and the genes that encode them affect the initial pathways of SSRI action. A polymorphism in the upstream regulatory site for the SERT gene (SLC6A4) has been widely studied. This SERT polymorphism (serotonin transporter linked polymorphic region; 5-HTTLPR) involves the presence or absence of a 44 base-pair segment, which produces a long (L) or short (S) allele; a difference that can influence transcriptional activity (Heils et al 1997; Lesch 1998). 5-HTTLPR has been associated with susceptibility to depression (Caspi et al 2003), although there is considerable heterogeneity between studies (Lotrich et al 2001; Lotrich and Pollock 2004).
The S allele has also been associated with diminished response to several SSRIs as compared with the L allele in multiple studies (Arias et al 2000; Pollock et al 2000; Zanardi et al 2000; Rausch, Johnson, et al 2002; Yu et al 2002; Arias et al 2003), with two exceptions in Asian populations (Kim et al 2000; Yoshida, Iko, et al 2002). The S allele may also increase vulnerability to SSRI side effects (Mundo et al 2001; Murphy GM et al 2003b). While the general finding of worse outcome in SSRI-treated patients with the S allele has been well replicated, discrepant reporting in several of these studies makes it difficult to determine the effect size of this polymorphism. Among issues to be further clarified is the effect of 5-HTTLPR in different ethnic populations; linkage disequilibrium with other polymorphisms in different ethnic populations; the effect size in different age groups and at different doses of SSRIs; delineating which depressive symptoms and side effects are influenced; and determining how this polymorphism interacts with other polymorphisms. Moreover, the role of other SLC6A4 polymorphisms remains comparatively unexamined (Lesch 1998; Battersby et al 1999; Michaelovsky et al 1999; Nakamura et al 2000; Ito et al 2002).
In addition to SERT, monoamine oxidase A (MAOA), although not typically expressed in 5-HT neurons, also affects 5-HT levels (Saura et al 1996; Richards et al 1998). A functional polymorphism in the upstream regulatory region for MAOA (Sabol et al 1998) can influence response to SSRIs (Yoshida, Naito et al 2002; Yoshida et al 2003). As with 5-HTTLPR, the frequency and effect size may differ among ethnic groups (Yoshida, Naito et al 2002). Monoamine oxidase B (MAOB) is present in 5-HT neurons but is less effective in metabolizing 5-HT than MAOA (Saura et al 1996; Richards et al 1998). MAOB polymorphisms (Balciuniene et al 2002) have been examined for associations with Parkinson’s disease, but we are unaware of any published studies examining a role in affecting SSRI response.
Tryptophan hydroxylase (TPH) is the rate-limited enzyme in 5-HT synthesis. A single nucleotide polymorphism (SNP), A218C, may be part of a GATA transcription factor binding site, and has been associated with suicidal behavior in Caucasians (Rujescu et al 2003). Initial reports associated this polymorphism with the rate of response to SSRIs (Serretti et al 2001a; Serretti, Zanardi, Rossini, et al 2001), although this was not replicated in a smaller study with Japanese subjects (Yoshida, Naito, et al 2002). A second gene coding for tryptophan hydroxylase has been identified; however, whether it is expressed in the human brain remains controversial (Patel et al 2004). An additional enzyme, methylenetetrahydrofolate reductase (MTHFR), is involved in monoamine synthesis, and an MTHFR polymorphism (C677T) has been associated with major depression (Bjelland et al 2003).
There are also a number of pathways indirectly regulating synaptic 5-HT by influencing SERT. For example, protein kinase C (PKC) activation can decrease SERT activity (Qian et al 1997; Ramamoorthy et al 1998). Conversely, SERT co-exists in a complex with protein phosphatase 2A (PP2A), and dephosphorylation by PP2A can increase transporter activity at the cell surface (Bauman et al 2000). Phosphorylation by calcium/calmodulin-dependent protein kinase II (CAMKII) may also enhance SERT activity (Yura et al 1996), as can various tyrosine kinases (Helmeste and Tang 1995). Moreover, transcription of the SERT gene may be affected by both PKC and protein kinase A (PKA) (Mossner et al 2000). Interestingly, given that the half-life of SERT is about 3 to 4 days (Vicentic et al 1999), a new steady-state in SERT levels would take about 2 weeks.
Serotonin receptors
At least 14 genes coding for 5-HT receptors are known, although post-transcriptional processing can result in over 30 5-HT receptor subtypes (Raymond et al 2001). Genetic differences in receptor number (Bmax), binding (Kd), kinetics (Vmax and Km), and desensitization rates for some of these receptors may potentially affect SSRI action. To date, there are data suggesting the plausible involvement of at least seven specific 5-HT receptors in mediating SSRI effects.
One class of 5-HT receptors (including 5-HT1a and 5-HT1b) interacts primarily with inhibitory G-proteins to decrease adenylyl cyclase (AC) activity. The 5-HT1a receptor has been implicated in many SSRI effects (Dhaenen 2001; Bourin et al 2002; Blier and Ward 2003; Monaca et al 2003). For example, blockade of 5-HT1a with antagonists or the use of 5-HT1a knockout mice prevents the effect of SSRIs on paradoxical sleep (Monaca et al 2003). Blockade of this receptor also can augment antidepressant response to SSRIs (Artigas et al 2001). Moreover, a 5-HT1a polymorphism (C-1019G) has recently been associated with vulnerability to major depression (Lemonde et al 2003). The 5-HT1b receptor has also been associated with irritability, aggression, impulsivity, and possibly mood disorders (Sanders et al 2002). In knockout mice lacking 5-HT1b, some effects of SSRIs are abolished (Monaca et al 2003), and 5-HT1b antagonists can also attenuate some SSRI effects (Millan et al 2003). Polymorphisms in the gene for 5-HT1b, including a common A(-161)T SNP in the promoter region, have been preliminarily associated with several psychiatric phenotypes, including suicide (Hawi et al 2002; Sanders et al 2002).
A second class of receptors includes 5-HT6 and 7. Each of these receptors primarily interacts with stimulatory G-proteins to increase adenylyl cyclase activity. Their function has been less studied to date. The 5-HT6 receptor is involved in neuronal serotonergic transmission and may have effects on anxiety and mood (Yoshioka et al 1998). Further, a 5-HT6 polymorphism (C267T) has been associated with Parkinson’s disease (Messina et al 2002). Although an initial study with 34 patients did not find an effect of a single 5-HT6 polymorphism on treatment response (Wu et al 2001), a more definitive study may be required to determine its potential involvement in SSRI efficacy. The 5-HT7 receptor is involved in hippocampal function (Gill et al 2002), and has been implicated in the regulation of the glucocorticoid receptor (Laplante et al 2002).
5-HT2a and 5-HT2c receptors primarily act at G-proteins associated with phospholipase C (PLC). The 5-HT2a receptor may play a role in SSRI response (Bonhomme and Esposito 1998). In fact, antagonism at 5-HT2a can augment the efficacy of SSRIs (Marek et al 2003), and response to SSRI treatment has been associated with normalization of 5-HT2a receptor binding (Messa et al 2003). Consistent with these findings, polymorphisms in the 5-HT2a gene (eg, G-1438A and C102T) may be associated with vulnerability to depression and suicide (Enoch et al 1999; Du et al 2000; Arias et al 2001; Du et al 2001; Holmes et al 2003), although this association has not been replicated in all studies (Bondy et al 2000; Minov et al 2001). Further, a 5-HT2a polymorphism (G-1438A) may be associated with the response to SSRIs in several populations (Cusin et al 2002; Sato et al 2002; Sugie et al 2003). A recent study in elderly depressed subjects suggested that a 5-HT2a polymorphism (C102T) may also be associated with sensitivity to the adverse effects of mirtazapine, but was without influence on SSRI adverse effects (Murphy GM et al 2003b).
SSRIs also affect the function of the 5-HT2c receptor (Bristow et al 2000), with some adverse effects potentially mediated by 5-HT2c. For example, the acute increases in anxiety reported with SSRIs may be attenuated with 5-HT2c antagonists, as assessed in animal models (Dekeyne et al 2000; Bagdy et al 2001). The 5-HT3 receptor, which is an ionophore, may also be involved in mood disorders (Niesler et al 2001) and sensitivity to some SSRI effects (Redrobe and Bourin 1997). For example, 5-HT3 antagonists can decrease SSRI-induced nausea (Wilde and Markham 1996).
G-proteins
Other than for the 5-HT3 receptor, most of the downstream effects of 5-HT are mediated by G-proteins (Raymond et al 2001). There are 16 genes for G-protein α subunits, 5 for β, and 12 for γ (Downes and Gautam 1999). In general, the αs family activates adenylyl cyclases, the αi family inhibits adenylyl cyclases, and the αq family activates phospholipase C isozymes. The βγ complex modulates the activity of the G protein α subunit. Many of these subunits are expressed within the brain (Downes and Gautam 1999).
The αs subunit, which mediates many of the effects of 5-HT6 and 5-HT7 receptors, has a polymorphism in exon 5 that has been associated with major depression (Zill et al 2002). As this subunit is expressed in many areas of the body, this polymorphism has also been associated with other disorders including hypertension, which may be comorbid with major depression (Jia et al 1999; Abe et al 2002). The expression of αs is increased by SSRI treatment (Lesch et al 1992). Polymorphisms in the G protein β3 subunit, which is expressed in the brain, have also been associated with both depression and cardiovascular disease (Bondy et al 2002; Zill et al 2002; Nurnberger et al 2003). Moreover, preliminary evidence supports a role for this polymorphism (C825T) in response to antidepressant treatment (Zill et al 2002; Serretti et al 2003), including SSRIs. The αi subunits, which mediate the effects of 5-HT1 receptors, have several isoforms expressed in the brain (Downes and Gautam 1999) and have been implicated in antidepressant response in mouse models (Galeotti et al 2002).
Adenylyl cyclases, protein kinases, phospholipases, and phosphodiesterases
Both the αs and αi G protein subunits directly affect the function of adenylyl cyclase (AC), resulting in either increased or decreased cyclic adenosine monophosphate (cAMP), respectively. There are at least 9 genes encoding 9 different AC enzymes. Most are expressed in the brain, although subtypes 1, 2, 6, 8, and 9 are more predominantly so (Defer et al 2000). Antidepressant treatment, including SSRIs, has been associated with increased transcription of AC1 (Jensen et al 2000). 5-HT1a activation decreases activity of neuronal AC2 (Albert et al 1999), and mice lacking AC8 exhibit altered stress-induced anxiety (Schaefer et al 2000). Each of these varied findings hints at a role for various AC isozymes in mediating the antidepressant effects of SSRIs. Initial studies of AC7 and AC9 polymorphisms have been performed, although these particular isoforms did not demonstrate an association with depression (Hellevuo et al 1997; Toyota et al 2002).
Adenylyl cyclase activity increases cAMP, which then binds to PKA. PKA is a tetramer containing two regulatory (R) subunits and two catalytic (C) subunits. Type II PKA is predominantly expressed in the brain. Activation by cAMP results in dissociation of the R and C subunits, allowing the C subunit to phosphorylate multiple substrates – including but not limited to the b-adrenergic receptor, the nicotinic acetylcholine receptor, sodium and calcium channels, N-methyl-D-aspartate (NMDA) receptor subunits, tyrosine hydroxylase, neurofilaments, microtubule associated protein 2 (MAP2), synapsin I, rabphilin, GABA A receptor subunits, SERT, and cAMP response element-binding protein (CREB) (Nibuya et al 1996; Wischmeyer and Karschin 1996). Abnormalities in PKA/CREB signaling have been associated with mood disorders (Shelton et al 1996; Rahman et al 1997; Chang et al 2003; Kohen et al 2003). For example, an increase in phosphorylated CREB was noted in the postmortem prefrontal cortexes of depressed victims of suicide, but only in antidepressant-free subjects (Odagaki et al 2001). Decreased CREB was observed, however, in the temporal cortexes of subjects who had had major depression, but only in antidepressant-free subjects (Dowlatshahi et al 1998). Although the relationship between PKA/CREB and major depression may be complex, antidepressants normalized the CREB measures in both studies.
Stimulated by these observations, several theories of SSRI action have hypothesized a primary role for CREB phosphorylation by PKA (Duman et al 1997; Shelton 2000), as reviewed below. However, other phosphorylation events may also be important. For example, in neurons, PKA RIIb (regulatory subunit IIb) mediates association with membranes, where PKA is coupled with MAP2. As noted above, PKA can phosphorylate MAP2, decreasing micro-tubule assembly, and potentially affecting dendritic and synaptic growth (Nestler et al 1989; Miyamoto et al 1997).
Chronic SSRI treatment is capable of increasing PKA in select brain areas (Perez et al 1991; Rausch, Gillespie, et al 2002) while decreasing PKA in others (Rosin et al 1995). Additionally, SSRI treatment can result in a translocation of PKA into various intracellular compartments, potentially corresponding to its eventual antidepressant effect (Mori et al 1998). Although abnormalities in PKA activation have been associated with differences in response to noradrenergic antidepressants (Gurguis et al 1999), a systematic study of polymorphisms affecting PKA has not yet been performed. Given the widespread expression of PKA subunits, polymorphisms that drastically affect function may be expected to have widespread deleterious effects (Stergiopoulos and Stratakis 2003).
The cAMP produced by adenylyl cyclase is recycled by phosphodiesterases (PDEs). A limited number of the multiple PDE isoforms are expressed in the brain. In particular, PDE4 exists in brain regions involved in mood (Cherry and Davis 1999). Interestingly, PDE4 inhibitors such as rolipram have antidepressant effects. Antidepressants affect transcription of PDE4 (Ye et al 2000; Miro et al 2002; Zhao et al 2003), and PDE4D may mediate some anti-depressant effects of rolipram in animal models (Zhang et al 2002).
As noted, CREB is directly activated by PKA. It is also notable that the chromosomal region containing CREB has been linked with depression (Zubenko et al 2002). CREB is a member of the leucine zipper family of transcription factors, activating or repressing multiple genes with the cAMP response element (CRE) promoter (Yamamoto et al 1988). Chronic antidepressant treatment, including SSRIs, can increase CREB mRNA in some brain areas (Nibuya et al 1996), and alterations in CREB activity have been associated with antidepressant effects. For example, increased peripheral CREB phosphorylation at two weeks was correlated with antidepressant response (Koch et al 2002). Conversely, increased CREB expression in the nucleus accumbens may adversely affect motivated behavior (Pliakas et al 2001; Newton et al 2002). CREB also increases BCL-2 (B-cell leukemia-2) expression (Riccio et al 1999) and is necessary for the increased brain-derived neurotrophic factor (BDNF). Both effects potentially influence neuronal survival (Conti et al 2002). Many genes have CRE in their promoter regions including nerve growth factor1A, corticotropin-releasing hormone, BDNF, and glucocorticoid and mineralocorticoid receptors.
Another protein that has recently been implicated in mediating the effects of SSRIs through PKA and other transduction pathways is the dopamine and cAMP regulated phosphoprotein (DARPP-32). DARPP-32 is an inhibitor of protein phosphatase-1 (PP1). Inhibiting protein phosphatase activity may amplify the effects of PKA. The efficacy of fluoxetine, an SSRI, is diminished in mice that are lacking DARPP-32 (Svenningsson et al 2002). Other protein phosphatases that may affect SSRI response will be reviewed below.
In addition to these effects immediately downstream from adenylyl cyclase activity, 5-HT activates PLC by 5-HT2 receptor-associated G-proteins. There are multiple isoforms for PLC. PLC, via creation of diacylglyceral, then activates PKC. Both PKC and PLC have been associated with depression (Shelton 2000), and there is evidence for PLCs involvement in mediating some SSRI effects (Rasenick et al 1996).
Polymorphisms in PLCγ1, such as an intronic dinucleotide repeat, have been associated with bipolar disorder and response to lithium treatment (Turecki et al 1998; Lovlie et al 2001). Additionally, antidepressant treatment can decrease the expression of PLCβ in animal models (Dwivedi et al 2002). 5-HT2-coupled G-proteins also activate phospholipase A2, releasing arachidonic acid from phospholipids. Via this pathway, SSRIs have been shown to increase the activity of phospholipase A2 (Qu et al 2003a, 2003b), and an intronic trinucleotide repeat polymorphism in phospholipase A2 may be associated with mood disorders (Meira-Lima et al 2003; Papadimitriou et al 2003).
PKC has at least 12 isoforms (Way et al 2000). In the brain, the isoforms α, β1,β2, γ, ε, δ, and ζ are expressed, although γ is most predominantly specific to the central nervous system (Saito and Shirai 2002). On activation, PKC translocates from the membrane to specific compartments such as the nucleus, cytoskeleton, and postsynaptic densities. Subsequent PKC substrates include vinculin, annexins, neuromodulin, and myrisoylated alanine-rich C-kinase substrate (MARCKS). PKC also desensitizes 5-HT2A receptors (Rahimian and Hrdina 1995) and downregulates SERT (Ramamoorthy et al 1998). Other neuronal functions include modulation of ion channels, norepinephrine transporters, and long-term potentiation (Saito and Shirai 2002). Interestingly, chronic treatment with SSRIs decreases PKC function in the cortex and hippocampus (Mann et al 1995).
Another effect of PLC is to increase the metabolism of phosphoinositol via activation of inositol phosphatase, with subsequent effects on cytoplasmic calcium levels. In turn, the increased calcium can activate CAMKII, which is the most abundant protein kinase in the brain and is particularly enriched at synaptic sites. Polymorphisms in some CAMKII isoforms have been demonstrated (Gloyn et al 2002), and both the α and β isoforms for CAMKII are expressed in the brain (Chang et al 2001). At the cellular level, CAMKII activation is associated with increased neurotransmitter release and long-term potentiation. For example, phosphorylation of the N-type calcium channel by CAMKII affects its interaction with SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins in influencing neurotransmitter release (Yokoyama et al 1997). Increased neurotransmitter release may also be affected by phosphorylation of synaptotagmin, a crucial element of calcium-induced exocytosis. Chronic antidepressant treatment increases CAMKII in selective brain areas (Popoli et al 1995; Pilc et al 1999; Consogno et al 2001; Celano et al 2003).
Protein phosphatases
Multiple protein phosphatases (PPs) are present in the brain to dephosphorylate proteins, reversing the effect of kinases (Price and Mumbly 1999). One example is PP1 which has at least three isoforms expressed in the brain (da Cruz e Silva et al 1995). Interaction with spinophilin results in localization of PP1 to the dendritic spines, while interaction with tau bridges PP1 to microtubules (Allen et al 1997). In dendritic spines, PP1 can dephosphorylate CAMKII and glutamate receptors (Kennedy 1998), and PP1 activity may be necessary for effects of 5-HT in acutely affecting prefrontal cortical neuronal activity (Cai et al 2002). There are also genes for several associated regulatory proteins. For example, DARRP-32, whose involvement in potentially affecting SSRI response was discussed above, is a regulator of PP1 (Svenningsson et al 2002).
A second phosphatase, PP2A, exists as a heterotrimer, with a catalytic, regulatory, and scaffolding subunit. Both the catalytic and scaffolding subunits exist throughout the brain, and there are several region-specific regulatory subunits (Stack et al 1998). In addition to other activities, PP2A can exist in close relationship with the SERT protein and is involved in regulating SERT’s activity (Bauman et al 2000). PP2B, also known as calcineurin, exists throughout the brain but is particularly enriched in hippocampal dendritic spines. It is activated by calcium/calmodulin (Klee et al 1998). Through the calcium/calmodulin/PP2B pathway, 5-HT2 receptor activation can inhibit calcium channels in the prefrontal cortex (Day et al 2002). However, along with several other PPs in the brain (PP2C, PP4, PP5, PP6) (Cohen 1997), a function in potentially modulating SSRI-induced intracellular transduction remains to be determined.
Regulatory and scaffolding proteins
Notably many of these intracellular transduction pathways interact in complex ways, depending on the intracellular compartment and association of other regulatory proteins (Sim and Scott 1999; Robinson-White and Stratakis 2002). In addition to the proteins discussed above, others may indirectly play roles. There are multiple regulators of G-protein signaling (RGS) (Chidiac and Roy 2003) that directly modulate G-protein function; multiple receptors for activated PKC (RACKs) that may be important for translocation of PKC to specific intracellular locations; and multiple anchoring proteins that can regulate the location, function, and interaction of various protein kinases (Scott 1997; Bayer and Schulman 2001). As just one example, A-kinase anchoring protein 79/150 (AKAP79/150) is an anchoring protein that interacts with PKA, PKC, and PP2B. It can recruit these three enzymes to glutamate receptors in the dendritic spine via an interaction with membrane-associated guanylate kinase scaffold proteins. Conversely, the activity of AKAP79/150 is regulated by PKC and calmodulin (Gomez et al 2002).
To briefly review, there is accumulating evidence in support of several protein kinases, protein phosphatases, and other related second messengers in mediating the effects of SSRIs. Importantly, there is also accumulating initial evidence in support of proximal, intracellular pathway polymorphisms affecting SSRI efficacy. However, for most of these proteins, the association of genetic variation and variable response to SSRIs has yet to be fully investigated.
Moreover, chronic administration of SSRIs can also affect various systems that are downstream from these intracellular transduction pathways. Several of these systems have been implicated in depression, as briefly outlined in the next sections, and may offer biologically plausible candidates for association analyses of SSRI response.
Hypothalamic-pituitary-adrenal axis
Stress and dysregulation of the hypothalamic-pituitary-adrenal (HPA) axis has long been associated with depression (Gold et al 1988; Gold and Chrousos 2002; Strohle and Holsboer 2003), and CRH antagonists may have antidepressant properties (Kunzel et al 2003). Because antidepressants can increase glucocorticoid receptor (GR) expression and potentially normalize the HPA axis, this has been proposed as one mechanism of action (Barden et al 1995; Holsboer 2000; Herr et al 2003). Antidepressants also can alter the synthesis and release of CRH, thereby normalizing the HPA axis (Brady et al 1991; Torres et al 1998). Consistent with this, polymorphisms in the CRH gene that affect cortisol secretion (Rosmond et al 2001) have been described (Gonzalez-Gay et al 2003), and may be associated with major depression (Claes et al 2003) as assessed using haplotype analyses.
An initial investigation of a CRH receptor 2 polymorphism (G1047A) also hinted at a possible role in mood disorders (Villafuerte et al 2002). Polymorphisms in GR have been associated with obesity (Di Blasio et al 2003), sensitivity to dexamethasone challenges (Huizenga et al 1998), and possibly to abnormal responses to stress (DeRijk et al 2002). Of note, mineralocorticoid receptor (MR) gene expression is also affected by chronic antidepressant treatment in rats (Brady et al 1991), and MR function has also been linked to the pathophysiology of depression (Young et al 2003).
Brain-derived neurotrophic factor and neurogenesis
SSRIs can increase neurogenesis in the hippocampus (Malberg et al 2000; Santarelli et al 2003), and hippocampal neurogenesis has been implicated as a requirement for the behavioral effects of antidepressants (Santarelli et al 2003). Increased BDNF, a neurotropin, may mediate this antidepressant effect of SSRIs (Nibuya et al 1995; Duman et al 1997; Altar 1999; Duman et al 1999; Shirayama et al 2002). BDNF may also, in turn, regulate the expression of SERT, an affect that can be modulated by the 5-HTTLPR (Mossner et al 2000). A polymorphism in BDNF has been preliminarily associated with bipolar disorder (Neves-Pereira et al 2002), although this was not found in a Chinese population of subjects with mood disorders (Hong et al 2003).
The receptor for BDNF is trkB (troponin/receptor kinase B), which is affected by antidepressant treatment, including SSRIs, and appears to be necessary for antidepressant effects (Rumajogee et al 2002; Saarelainen et al 2003). Moreover, antidepressants not only increase the expression of BDNF and trkB, but also neurotrophin-3, (Nibuya et al 1995; Smith et al 1995; Nibuya et al 1996; Duman et al 1997). Of note, transcription for each of these three is increased by CREB (Duman et al 1997). The cAMP/PKA/CREB mediated effects may therefore be ultimately mediated by BDNF acting at the trkB receptor (Galter and Unsicker 2000).
Several intracellular kinases transduce BDNF effects. For example, there are mitogen-activated protein (MAP) kinase cascades, which also involve extracellular signal-regulated protein kinase (ERK) (Gould and Manji 2002; Manji and Chen 2002). BDNF infusion increases ERK phosphorylation and immediate early genes (Shirayama et al 2002). Inhibition of mitogen and extracellular signal-regulated protein kinase (MEK), which mediates the BNDF-induced phosphorylation of ERK, prevents the anti-depressant effects of BDNF in animal models (Shirayama et al 2002). Both MAP kinase and ERK have been implicated in mediating the 5-HT1A effect of inhibiting caspase-3 and apoptosis (Adayev et al 2003). Abnormalities in ERK1 and ERK2 have been noted in animal models of depression (Feng et al 2003), and decreased neuronal ERK expression is seen in victims of suicide (Dwivedi et al 2001). Other potential downstream effector molecules for BDNF that may enhance neuronal survival, augment neuroplasticity, or decrease apoptosis include Bcl-2 and glycogen synthase kinase 3 (Manji et al 2001; Gould and Manji 2002; Manji and Chen 2002).
Additional neurotransmitters
There are multiple neurotransmitter systems that interact in complex ways. Further, many of these neurotransmitter systems share the same intracellular transduction pathways. The cholinergic, noradrenergic, and dopaminergic systems may all be dysregulated in depression (Leonard 1996; Chau et al 1999; Iversen 2000; Schatzberg et al 2002; Shytle et al 2002; Shaffery et al 2003). In fact, various polymorphisms in the norepinephrine and dopamine systems have been associated with mood disorders (Jonsson et al 1998; Mason et al 1999; Mitchell et al 2000; Suzuki et al 2001; Massat et al 2002; Ni et al 2002; Ranade et al 2002; Rosmond et al 2002; Urwin et al 2002; Jonsson et al 2003; Ueno 2003). A polymorphism in the muscarinic-2 receptor has also been associated with depression (Comings et al 2002). Preliminary studies do not support roles for select polymorphisms in D2 and D4 dopamine receptors in affecting treatment response (Serretti et al 2001b). However, polymorphisms in other monaminergic and cholinergic receptors may interact with SSRI efficacy. For example, catachol-O-methyltransferase primarily metabolizes dopamine and norepinephrine and has a polymorphism that has been associated with poor response to SSRIs (Catalano 2001).
There are abnormalities in the glutamatergic system in depression (Skolnick 2002; Sanacora et al 2003). Moreover, enhancement of α-amino-3-hydroxy-5-methyl-4-isoxazole proprionic acid (AMPA) receptor activity and decreased NMDA receptor activity may potentiate the antidepressant effect of SSRIs (Rogoz et al 2002; Li et al 2003). Of the 5 genes coding for NMDA receptor subunits (NR1 and NR2A-NR2D) and 4 for AMPA receptors (GluR1-GluR4), a polymorphism in the NR1 gene has preliminarily been associated with mood disorders (Mundo et al 2003).
The nitric oxide system also interacts with 5-HT in moderating behavior (McLeod et al 2001; Chiavegatto and Nelson 2003). Interestingly, a polymorphism for neuronal nitric oxide synthetase (C29T) has been preliminarily associated with depression and response to SSRIs (Yu, Chen, Wang, et al 2003). Vasopressin (Keck et al 2003), neuropeptide Y (Caberlotto et al 1998; Pandey et al 2003), and dynorphin (Chen et al 2002; Newton et al 2002) may also play roles.
Ion channels and synaptic transmission
As noted above, 5-HT transmission and subsequent intracellular transduction cascades can also effect ion channels, thereby affecting neuronal excitability and neurotransmitter release (Nunoki et al 1989; Blier and de Montigny 1994; Levitan 1994; Wischmeyer and Karschin 1996; Fairchild et al 2003). One example is the G-protein-gated inwardly rectifying potassium channel (GIRK), which regulates membrane excitability. At least three subunits for GIRK are native to hippocampal neurons, Kir3.1, Kir3.2A, and Kir3.2C, and these may mediate some 5-HT1A responses (Fairchild et al 2003).
SSRIs can also decrease synaptic function in select areas by influencing calcium channels. For example, inhibition of P/Q-type calcium channels in cerebrocortical neurons by fluoxetine can be mediated by a PKC pathway, resulting in decreased neurotransmission (Wang et al 2003). Consistent with these observations, blockade of calcium channels using verapamil or diltiazem can attenuate the effectiveness of SSRIs in animal models of depression (Srivastava and Nath 2000).
Other molecules affecting synaptic transmission have been similarly implicated in the downstream effects of antidepressants. For example Pyk2 (proline-rich tyrosine kinase 2) is a tyrosine kinase that can modify the activity of NMDA receptors and voltage-gated potassium channels in the lateral septum, enhancing synaptic transmission and long-term potentiation (Sheehan et al 2003). Other potential moderators of antidepressant response include apolipoprotein E (ApoE) and various immunologic cytokines that may affect depression (Song et al 1998; Alvarez et al 2002; Anisman et al 2002; Anisman and Merali 2002; Murphy GM et al 2003a; Yu, Chen, Hong, et al 2003).
Unanticipated candidates based on microarray and genomic screening
A recent microarray study examining whole frontal cortex (Landgrebe et al 2002) has identified 16 genes that were influenced by SSRI treatment in mice – all were down-regulated. Only one related to signal transduction, six were involved in metabolism, two in cell-cycle regulation, and one was a homeobox transcription factor. An alternative approach has been to examine the effect of chronic stress in tree shrews, using a subtractive hippocampal cDNA library. Chronic stress decreased the expression of neural growth factor, membrane glycoprotein 6a, CDC-like kinase 1, and G-protein subunit αq. Treatment with clomipramine mitigated these transcriptome changes (Alfonso et al 2004). If confirmed, the findings from this nascent technology may eventually provide unsuspected, novel candidate genes for association analyses.
Summary
A number of neurotransmitter and neuromodulatory systems are associated both with mood disorders such as MDD and response to antidepressant treatment. Each of these systems offers a biologically plausible target for selection of candidate genes affecting SSRI response. Empirically delineating the genetic polymorphisms in these various systems that, in fact, influence SSRI response will be an important goal for the next generation of studies examining treatment outcome.
Given that many of the proteins, such as adenylyl cyclase, Gi subunits, and protein phosphatases have multiple isoforms, selecting specific isoforms for genetic analysis may require first determining which isoforms are involved in neuronal SSRI-influenced pathways. Further, once a specific gene is chosen for analysis, the next step requires selecting which polymorphism(s) within the gene to study. Because genes often have multiple polymorphisms, it is unfeasible to assess them all. Conversely, studies that test only a single polymorphism may erroneously conclude that a gene is not involved in SSRI response, but miss testing an important functional polymorphism in that gene. Often, it is ideal to choose polymorphisms with known functional affects. It may be useful to choose polymorphisms that have had previously demonstrated associations with other phenotypes of interest such as MDD. Ultimately, selecting a subset of several polymorphisms within a gene for haplotype analysis may provide improved power over choosing only a single polymorphism for study (Hoehe et al 2003). This is of particular relevance for interpreting those studies that report a lack of association for single polymorphisms. If the complete gene including its regulatory regions is not examined, functionally important genetic differences may be missed and lack of importance for a gene erroneously inferred. This is a critical shortcoming in many recent papers reporting “negative” findings.
Discussion
As individual polymorphisms are likely to have small effect sizes, identifying multiple genes may be necessary to generate a complete picture of genetic variability and responsiveness to SSRIs. By identifying likely targets based on current knowledge regarding the mechanisms of SSRI action, a variety of potential candidates have been culled. This review has attempted to suggest multiple candidate genes based on known and putative mechanisms of SSRI action. Notably, the number of targeted genes represents less than 0.5% of the genome. With current high-throughput methodologies for assessing polymorphisms, examining this large but significantly limited number of genes is feasible (Cowan et al 2002).
However, studies of variable antidepressant response are affected by several methodologic issues that are inherent in antidepressant trials. There are fundamental considerations regarding the phenotypic definition of antidepressant response (Nierenberg 2003). For example, although depression is a complex and polythetic disorder (Kendler et al 1992; Kendell 2001), it is often treated as a single entity. Similarly, defining response is problematic (Nierenberg and DeCecco 2001). Response can be quantified as the rate of response or the magnitude of response. It can also be qualitatively defined as achieving a predetermined level of symptomatology. Different genes may specifically affect different aspects and/or subtypes of depression, or they may specifically affect different elements of response. That is, there may be a number of “mood disorders”, or even subtypes of “major depression”, that may respond differently to SSRIs. Additionally, some genetic variation may be associated with different types of side effects and different rates of developing tolerance to side effects, rather than antidepressant response itself (Murphy GM et al 2003b). Unfortunately, there is extremely little data to date regarding the association of polymorphisms with different measures of response, tolerance to SSRIs, or different side effects of SSRIs.
Moreover, determining true drug responders is also complicated by partial or non-adherence to the medication (Thompson et al 2000). In addition to variable adherence, the concentration of SSRIs can be influenced by dosage and environmental factors. The effect of some polymorphisms may be evident at low SSRI concentrations but not at higher concentrations, or vice versa. For example, we have noted that 5-HTTLPR is associated with variable response to paroxetine in the first few weeks, but that response to the SSRI did not differ after several weeks (Pollock et al 2000). Given that paroxetine can inhibit its own metabolism, we observed that paroxetine levels increased after several weeks in elderly individuals, to concentrations potentially sufficient for noradrenergic involvement (Gilmor et al 2002). This is of interest given that 5-HTTLPR was not associated with response to a noradrenergic reuptake inhibitor (Pollock et al 2000). For this reason, as well as statistical reasons, accounting for adherence and SSRI exposure may be essential in association studies of treatment response (Judson 2003; Selinger-Leneman et al 2003). Accounting for adherence and SSRI exposure in these types of studies has been extremely limited to date.
There is also the ever-escalating problem of placebo response rates in clinical trials (Walsh et al 2002). This is complicated by the aggregation of potential subtypes of depression in clinical studies and the relative exclusion of other subtypes, with some subtypes possibly having variable drug responsiveness (Parker et al 2002). If these confounding elements are not accounted for, erroneous conclusions are possible.
Population stratification is another concern. Methods such as genomic control and structured association may be used to correct for population substructuring (Devlin et al 2001). However, ethnic and cultural difference may confound results in other ways (Balant and Balant-Gorgia 2000). For example, adherence, social background, and ethnic differences may all interact to affect treatment response (Lotrich et al 2003), and the frequency of many polymorphisms varies with ancestry. Given these methods are now available for accounting for population stratification (eg, Devlin et al 2001), they should be included in these types of association studies.
There are also statistical issues pertaining to testing multiple polymorphisms. More sophisticated modeling may be necessary to accurately determine appropriate significance levels when testing multiple associations (Lucek et al 1998). For example, one suggested approach is to control for the false discovery rate (FDR), rather than focus on controlling the rate of false positives or false negatives (van den Oord and Sullivan 2003). That is, rather than stringently protecting against a single false rejection of a null hypothesis, FDR limits the percentage of false rejections, providing improved power when testing many likely candidate hypotheses (Devlin et al 2003). Further, there are additional methodological issues such as appropriate sample sizes (McCarthy and Hilfiker 2000) and genotyping procedures (Schulze et al 2003) that require consideration. Many studies reviewed above have insufficient sample sizes to meaningfully interpret “negative” findings.
Future studies carefully attending to these issues will likely yield a useful picture regarding the genetic variation affecting SSRI response. Candidate genes for these association analyses, selected from known and putative pathways of SSRI action, include polymorphisms affecting the serotonin transporter, serotonin receptors, intracellular transduction, the HPA axis, BDNF and neurogenesis, and other neurotransmitter systems. A complete picture of genetic variation will involve determining the relative role of multiple polymorphisms, their effect sizes, their interactions, their interactions with pharmacokinetic differences, and their relationship with environmental factors that influence treatment outcome.
As noted above, there are many methodological issues that require close consideration. There is wide variation among studies in attention to these issues (eg, careful definition of the study population in terms of ethnicity, demographics, environment, diagnoses, and comorbidities; careful definition of types of response; controlling for genetic variability in placebo response; controlling for differences in medication exposure; appropriate statistical analyses and attention to population substructure; and appropriate selection of a set of polymorphisms across the candidate gene).
This is a nascent, but quickly maturing field. To date, studies that have carefully attended to these concerns are very limited. Nonetheless, it is reasonable to predict that the goal of genetically determining which individual patients will benefit from SSRIs and which should be targeted for alternative therapies may be attainable in the near future.
Acknowledgments
Supported by NIMH grants MH65416, MH30915, MH52247, and MH16804.
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole proprionic acid
- AKAP79/150
A-kinase anchoring protein 79/150
- AC
adenylyl cyclase
- ApoE
apolipoprotein E
- BCL-2
B-cell leukemia-2
- GluR1-GluR4
AMPA receptor subunits
- BDNF
brain-derived neurotrophic factor
- CAMKII
calcium/calmodulin-dependent protein kinase II
- CRH
corticotropin-releasing hormone
- cAMP
cyclic adenosine monophosphate
- CRE
cAMP response element
- CREB
cAMP response element-binding protein
- DARPP-32
dopamine and cAMP regulated phosphoprotein
- ERK
extracellular signal-regulated protein kinase
- GIRK
G-protein-gated inwardly rectifying potassium channel
- Kir
GIRK subunit
- GR
glucocorticoid receptor
- HPA
hypothalamic-pituitary-adrenal
- MDD
major depressive disorder
- MTHFR
methylenetetrahydrofolate reductase
- MAP2
microtubule associated protein 2
- MR
mineralocorticoid receptor
- MAP
mitogen-activated protein
- MEK
mitogen and extracellular signal-regulated protein kinase
- MAOA
monoamine oxidase A
- MAOB
monoamine oxidase B
- MARCKS
myrisoylated alanine-rich C-kinase substrate
- NMDA
N-methyl-D-aspartate
- NR
NMDA receptor subunit
- PDE
phosphodiesterase
- PLC
phospholipase C
- Pyk2
proline-rich tyrosine kinase 2
- PKA
protein kinase A
- PKA RIIb
PKA regulatory subunit IIb
- PKC
protein kinase C
- PP
protein phosphatase
- RACK
receptor for activated PKC
- RGS
regulator of G-protein signaling
- SSRI
selective serotonin reuptake inhibitor
- 5-HT
serotonin
- SERT
serotonin transporter
- SLC6A4
SERT gene
- 5-HTTLPR
serotonin transporter linked polymorphic region
- SNP
single nucleotide polymorphism
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- trkB
troponin/receptor kinase B
- TPH
tryptophan hydroxylase
References
- Abe M, Nakura J, Yamamoto M, et al. Association of GNAS1 gene variant with hypertension depending on smoking status. Hypertension. 2002;40:261–5. doi: 10.1161/01.hyp.0000028490.77489.0c. [DOI] [PubMed] [Google Scholar]
- Adayev T, Ray I, Sondhi R, et al. The G protein-coupled 5-HT1A receptor causes suppression of caspase-3 through MAPK and protein kinase Calpha. Biochimica Biophys Acta. 2003;1640:85–96. doi: 10.1016/s0167-4889(03)00023-5. [DOI] [PubMed] [Google Scholar]
- Albert PR, Sajedi N, Lemonde S, et al. Constitutive G(i2)-dependent activation of adenylyl cyclase type II by the 5-HT1A receptor. Inhibition by anxiolytic partial agonists. J Biol Chem. 1999;274:35469–74. doi: 10.1074/jbc.274.50.35469. [DOI] [PubMed] [Google Scholar]
- Alfonso J, Pollevick GD, Van Der Hart MG, et al. Identification of genes regulated by chronic psychosocial stress and antidepressant treatment in the hippocampus. Eur J Neurosci. 2004;19:659–66. doi: 10.1111/j.1460-9568.2004.03178.x. [DOI] [PubMed] [Google Scholar]
- Allen PB, Ouimet CC, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci U S A. 1997;94:9956–61. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altar CA. Neurotrophins and depression. Trends Pharmacol Sci. 1999;20:59–61. doi: 10.1016/s0165-6147(99)01309-7. [DOI] [PubMed] [Google Scholar]
- Alvarez V, Mata IF, Gonzalez P, et al. Association between the TNFa-308 A/G polymorphism and the onset-age of Alzheimer disease. Am J Med Genet. 2002;114:574–7. doi: 10.1002/ajmg.10515. [DOI] [PubMed] [Google Scholar]
- Anisman H, Kokkinidis L, Merali Z. Further evidence for the depressive effects of cytokines: anhedonia and neurochemical changes. Brain Behav Immun. 2002;16:544–56. doi: 10.1016/s0889-1591(02)00011-9. [DOI] [PubMed] [Google Scholar]
- Anisman H, Merali Z. Cytokines, stress, and depressive illness. Brain Behav Immun. 2002;16:513–24. doi: 10.1016/s0889-1591(02)00009-0. [DOI] [PubMed] [Google Scholar]
- Arias B, Catalan R, Gasto C, et al. 5-HTTLPR polymorphism of the serotonin transporter gene predicts non-remission in major depression patients treated with citalopram in a 12-weeks follow up study. J Clin Psychopharmacol. 2003;23:563–7. doi: 10.1097/01.jcp.0000095350.32154.73. [DOI] [PubMed] [Google Scholar]
- Arias B, Gasto C, Catalan R, et al. Variation in the serotonin transporter gene and clinical response to citalopram in major depression. Am J Med Genet. 2000;96:536. [Google Scholar]
- Arias B, Gasto C, Catalan R, et al. The 5-HT(2A) receptor gene 102T/C polymorphism is associated with suicidal behavior in depressed patients. Am J Med Genet. 2001;105:801–4. doi: 10.1002/ajmg.10099. [DOI] [PubMed] [Google Scholar]
- Artigas F, Celada P, Laruelle M, et al. How does pindolol improve antidepressant action? Trends Pharmacol Sci. 2001;22:224–8. doi: 10.1016/s0165-6147(00)01682-5. [DOI] [PubMed] [Google Scholar]
- Bagdy G, Graf M, Anheuer Z, et al. Anxiety-like effects induced by acute fluoxetine, sertraline or m-CPP treatment are reversed by pretreatment with the 5-HT2C receptor antagonist SB-242084 but not the 5-HT1A receptor antagonist WAY-100635. Int J Neuropsychopharmacol. 2001;4:399–408. doi: 10.1017/S1461145701002632. [DOI] [PubMed] [Google Scholar]
- Balant LP, Balant-Gorgia EA. Cultural differences: implications on drug therapy and global drug development. Int J Clin Pharmacol Therap. 2000;38:47–52. doi: 10.5414/cpp38047. [DOI] [PubMed] [Google Scholar]
- Balciuniene J, Emilsson L, Oreland L, et al. Investigation of the functional effect of monoamine oxidase polymorphisms in human brain. Hum Genet. 2002;110:1–7. doi: 10.1007/s00439-001-0652-8. [DOI] [PubMed] [Google Scholar]
- Barden N, Reul JM, Holsboer F. Do antidepressants stabilize mood through actions on the hypothalamic-pituitary-adrenocortical system? Trends Neurosci. 1995;18:6–11. doi: 10.1016/0166-2236(95)93942-q. [DOI] [PubMed] [Google Scholar]
- Battersby S, Ogilvie AD, Blackwood DHR, et al. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3′ untranslated region of the human serotonin transporter gene. J Neurochem. 1999;72:1384–8. doi: 10.1046/j.1471-4159.1999.721384.x. [DOI] [PubMed] [Google Scholar]
- Bauman AL, Apparsundaram S, Ramamoorthy S, et al. Cocaine and antidepressant-sensitive biogenic amine transporters exist in regulated complexes with protein phosphatase 2A. J Neurosci. 2000;20:7571–8. doi: 10.1523/JNEUROSCI.20-20-07571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer KU, Schulman H. Regulation of signal transduction by protein targeting: the case for CaMKII. Biochem Biophys Res Commun. 2001;289:917–23. doi: 10.1006/bbrc.2001.6063. [DOI] [PubMed] [Google Scholar]
- Bjelland I, Tell GS, Vollset SE, et al. Folate, vitamin B12, homocysteine, and the MTHFR 677C->T polymorphism in anxiety and depression: the Hordaland Homocysteine Study. Arch Gen Psychiatry. 2003;60:618–26. doi: 10.1001/archpsyc.60.6.618. [DOI] [PubMed] [Google Scholar]
- Blier P, de Montigny C. Current advances and trends in the treatment of depression. Trends Pharmacol Sci. 1994;15:220–5. doi: 10.1016/0165-6147(94)90315-8. [DOI] [PubMed] [Google Scholar]
- Blier P, Ward NM. Is there a role for 5-HT1A agonists in the treatment of depression? Biol Psychiatry. 2003;53:193–203. doi: 10.1016/s0006-3223(02)01643-8. [DOI] [PubMed] [Google Scholar]
- Bondy B, Baghai TC, Zill P, et al. Combined action of the ACE D- and the G-protein beta3 T-allele in major depression: a possible link to cardiovascular disease? Mol Psychiatry. 2002;7:1120–6. doi: 10.1038/sj.mp.4001149. [DOI] [PubMed] [Google Scholar]
- Bondy B, Kuznik J, Baghai T, et al. Lack of association of serotonin-2A receptor gene polymorphism (T102C) with suicidal ideation and suicide. Am J Med Genet. 2000;96:831–5. doi: 10.1002/1096-8628(20001204)96:6<831::aid-ajmg27>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Bonhomme N, Esposito E. Involvement of serotonin and dopamine in the mechanism of action of novel antidepressant drugs: a review. J Clin Psychopharmacol. 1998;18:447–54. doi: 10.1097/00004714-199812000-00005. [DOI] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–36. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Bourin M, David DJ, Jolliet P, et al. Mechanism of action of antidepressants and therapeutic perspectives. Therapie. 2002;57:385–96. [PubMed] [Google Scholar]
- Brady LS, Whitfield HJJ, Fox RJ, et al. Long-term antidepressant administration alters corticotropin-releasing hormone, tyrosine hydroxylase, and mineralocorticoid receptor gene expression in rat brain: therapeutic implications. J Clin Invest. 1991;87:831–7. doi: 10.1172/JCI115086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow LJ, O’Connor D, Watts R, et al. Evidence for accelerated desensitisation of 5-HT(2C) receptors following combined treatment with fluoxetine and the 5-HT(1A) receptor antagonist, WAY 100,635, in the rat. Neuropharmacology. 2000;39:1222–36. doi: 10.1016/s0028-3908(99)00191-4. [DOI] [PubMed] [Google Scholar]
- Brosen K. The pharmacogenetics of the selective serotonin reuptake inhibitors. Clin Investig. 1993;71:1002–9. doi: 10.1007/BF00180032. [DOI] [PubMed] [Google Scholar]
- Buck KJ. Strategies for mapping and identifying quantitative trait loci specifying behavioral responses to alcohol. Alcohol Clin Exp Res. 1995;19:795–801. doi: 10.1111/j.1530-0277.1995.tb00949.x. [DOI] [PubMed] [Google Scholar]
- Buysse DJ. Insomnia, depression and aging. Assessing sleep and mood interactions in older adults. Geriatrics. 2004;59:47–51. [PubMed] [Google Scholar]
- Caberlotto L, Fuxe K, Overstreet DH, et al. Alterations in neuropeptide Y and Y1 receptor mRNA expression in brains from an animal model of depression: region specific adaptation after fluoxetine treatment. Brain Res Mol Brain Res. 1998;59:58–65. doi: 10.1016/s0169-328x(98)00137-5. [DOI] [PubMed] [Google Scholar]
- Cai X, Gu Z, Zhong P, et al. Serotonin 5-HT1A receptors regulate AMPA receptor channels through inhibiting Ca2+/calmodulin-dependent kinase II in prefrontal cortical pyramidal neurons. J Biol Chem. 2002;277:36553–62. doi: 10.1074/jbc.M203752200. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:368–89. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Catalano M. The challenges of psychopharmacogenetics. Am J Hum Genet. 1999;65:606–10. doi: 10.1086/302559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano M. Functionally gene-linked polymorphic regions and genetically controlled neurotransmitters metabolism. Eur Neuropsychopharmacol. 2001;11:431–9. doi: 10.1016/s0924-977x(01)00120-1. [DOI] [PubMed] [Google Scholar]
- Celano E, Tiraboschi E, Consogno E, et al. Selective regulation of presynaptic calcium/calmodulin-dependent protein kinase II by psychotropic drugs. Biol Psychiatry. 2003;53:442–9. doi: 10.1016/s0006-3223(02)01491-9. [DOI] [PubMed] [Google Scholar]
- Chang A, Li PP, Warsh JJ. cAMP-Dependent protein kinase (PKA) subunit mRNA levels in postmortem brain from patients with bipolar affective disorder (BD) Brain Res Mol Brain Res. 2003;116:27–37. doi: 10.1016/s0169-328x(03)00211-0. [DOI] [PubMed] [Google Scholar]
- Chang BH, Mukherji S, Soderling TR. Calcium/calmodulin-dependent protein kinase II inhibitor protein: localization of isoforms in rat brain. Neuroscience. 2001;102:767–77. doi: 10.1016/s0306-4522(00)00520-0. [DOI] [PubMed] [Google Scholar]
- Chau D, Rada PV, Kosloff RA, et al. Cholinergic, M1 receptors in the nucleus accumbens mediate behavioral depression. A possible downstream target for fluoxetine. Ann N Y Acad Sci. 1999;877:769–74. doi: 10.1111/j.1749-6632.1999.tb09320.x. [DOI] [PubMed] [Google Scholar]
- Chen AC, LaForge KS, Ho A, et al. Potentially functional polymorphism in the promoter region of prodynorphin gene may be associated with protection against cocaine dependence or abuse. Am J Med Genet. 2002;114:429–35. doi: 10.1002/ajmg.10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JA, Davis RL. Cyclic AMP phosphodiesterases are localized in regions of the mouse brain associated with reinforcement, movement, and affect. J Comp Neurol. 1999;407:287–301. [PubMed] [Google Scholar]
- Chiavegatto S, Nelson RJ. Interaction of nitric oxide and serotonin in aggressive behavior. Horm Behav. 2003;44:223–41. doi: 10.1016/j.yhbeh.2003.02.002. [DOI] [PubMed] [Google Scholar]
- Chidiac P, Roy AA. Activity, regulation, and intracellular localization of RGS proteins. Receptors Channels. 2003;9:135–47. [PubMed] [Google Scholar]
- Claes S, Villafuerte S, Forsgren T, et al. The corticotropin-releasing hormone binding protein is associated with major depression in a population from Northern Sweden. Biol Psychiatry. 2003;54:867–72. doi: 10.1016/s0006-3223(03)00425-6. [DOI] [PubMed] [Google Scholar]
- Cohen LJ, De Vane CL. Clinical implications of antidepressant pharmacokinetics and pharmacogenetics. Ann Pharmacother. 1996;30:1471–80. doi: 10.1177/106002809603001216. [DOI] [PubMed] [Google Scholar]
- Cohen PTW. Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem Sci. 1997;22:245–51. doi: 10.1016/s0968-0004(97)01060-8. [DOI] [PubMed] [Google Scholar]
- Comings DE, Wu S, Rostamkhani M, et al. Association of the muscarinic cholinergic 2 receptor (CHRM2) gene with major depression in women. Am J Med Genet. 2002;114:527–9. doi: 10.1002/ajmg.10406. [DOI] [PubMed] [Google Scholar]
- Consogno E, Racagni G, Popoli M. Modifications in brain CaM kinase II after long-term treatment with desmethylimipramine. Neuropsychopharmacology. 2001;24:21–30. doi: 10.1016/S0893-133X(00)00176-7. [DOI] [PubMed] [Google Scholar]
- Conti AC, Cryan JF, Dalvi A, et al. cAMP response element-binding protein is essential for the upregulation of brain-derived neurotrophic factor transcription, but not the behavioral or endocrine responses to antidepressant drugs. J Neurosci. 2002;22:3262–8. doi: 10.1523/JNEUROSCI.22-08-03262.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan WM, Kopnisky KL, Hyman SE. The human genome project and its impact on psychiatry. Annu Rev Neurosci. 2002;25:1–50. doi: 10.1146/annurev.neuro.25.112701.142853. [DOI] [PubMed] [Google Scholar]
- Cusin C, Serretti A, Zanardi R, et al. Influence of monoamine oxidase A and serotonin receptor 2A polymorphisms in SSRI antidepressant activity. Int J Neuropsychopharmacol. 2002;5:27–35. doi: 10.1017/S1461145701002711. [DOI] [PubMed] [Google Scholar]
- da Cruz e Silva EF, Fox CA, Ouiment CC, et al. Differential expression of protein phosphatase 1 isoforms in mammalian brain. J Neurosci. 1995;15:3375–89. doi: 10.1523/JNEUROSCI.15-05-03375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AK. Pharmacogenetics of the major polymorphic metabolizing enzymes. Fundam Clin Pharmacol. 2003;17:27–41. doi: 10.1046/j.1472-8206.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- Day M, Olson PA, Platzer J, et al. Stimulation of 5-HT(2) receptors in prefrontal pyramidal neurons inhibits Ca(v)1.2 L type Ca(2+) currents via a PLCbeta/IP3/calcineurin signaling cascade. J Neurophysiol. 2002;87:2490–504. doi: 10.1152/jn.00843.2001. [DOI] [PubMed] [Google Scholar]
- Defer N, Best-Belpomme M, Hanoune J. Tissue specificity and physiological relevance of various isoforms of adenylyl cyclase. Am J Physiol Renal Physiol. 2000;279:F400–16. doi: 10.1152/ajprenal.2000.279.3.F400. [DOI] [PubMed] [Google Scholar]
- Dekeyne A, Denorme B, Monneyron S, et al. Citalopram reduces social interaction in rats by activation of serotonin (5-HT)(2C) receptors. Neuropharmacology. 2000;39:1114–17. doi: 10.1016/s0028-3908(99)00268-3. [DOI] [PubMed] [Google Scholar]
- DeRijk RH, Schaaf M, de Kloet ER. Glucocorticoid receptor variants: clinical implications. J Steroid Biochem Mol Biol. 2002;81:103–22. doi: 10.1016/s0960-0760(02)00062-6. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Bacanu S-A. Unbiased methods for population-based association studies. Genet Epidemiol. 2001;21:273–84. doi: 10.1002/gepi.1034. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Analysis of multilocus models of association. Genet Epidemiol. 2003;25:36–47. doi: 10.1002/gepi.10237. [DOI] [PubMed] [Google Scholar]
- Dhaenen H. Imaging the serotonergic system in depression. Eur Arch Psychiatry Clin Neurosci. 2001;251:1176–80. [PubMed] [Google Scholar]
- Di Blasio AM, van Rossum EF, Maestrini S, et al. The relation between two polymorphisms in the glucocorticoid receptor gene and body mass index, blood pressure and cholesterol in obese patients. Clin Endocrinol. 2003;59:68–74. doi: 10.1046/j.1365-2265.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- Dowlatshahi D, MacQueen GM, Wang JF, et al. Increased temporal cortex CREB concentrations and antidepressant treatment in major depression. Lancet. 1998;352:1754–5. doi: 10.1016/S0140-6736(05)79827-5. [DOI] [PubMed] [Google Scholar]
- Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62:544–52. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- Drevets WC. Functional neuroimaging studies of depression: the anatomy of melancholia. Annu Rev Med. 1998;49:341–61. doi: 10.1146/annurev.med.49.1.341. [DOI] [PubMed] [Google Scholar]
- Du L, Bakish D, Lapierre YD, et al. Association of polymorphism of serotonin 2A receptor gene with suicidal ideation in major depressive disorder. Am J Med Genet. 2000;96:56–60. doi: 10.1002/(sici)1096-8628(20000207)96:1<56::aid-ajmg12>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Du L, Faludi G, Palkovits M, et al. Serotonergic genes and suicidality. Crisis J Crisis Interv Suicide. 2001;22:54–60. doi: 10.1027//0227-5910.22.2.54. [DOI] [PubMed] [Google Scholar]
- Duman RS, Heninger GR, Nestler EJ. A molecular and cellular theory of depression. Arch Gen Psychiatry. 1997;54:597–606. doi: 10.1001/archpsyc.1997.01830190015002. [DOI] [PubMed] [Google Scholar]
- Duman RS, Malberg JE, Thome J. Neural plasticity to stress and antidepressant treatment. Biol Psychiatry. 1999;46:1181–91. doi: 10.1016/s0006-3223(99)00177-8. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Agrawal A, Rizavi H, et al. Antidepressants reduce phosphoinositide-specific phospholipase C (PI-PLC) activity and the mRNA and protein expression of selective PLC beta 1 isozyme in rat brain. Neuropharmacology. 2002;43:1269–79. doi: 10.1016/s0028-3908(02)00253-8. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi H, Roberts RC, et al. Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. J Neurochem. 2001;77:916–28. doi: 10.1046/j.1471-4159.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Goldman D, Barnett R, et al. Association between seasonal affective disorder and the 5-HT2A promoter polymorphism, -1438G/A. Mol Psychiatry. 1999;4:89–92. doi: 10.1038/sj.mp.4000439. [DOI] [PubMed] [Google Scholar]
- Evans WE, McLeod HL. Pharmacogenomics – drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–49. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- Fairchild G, Leitch MM, Ingram CD. Acute and chronic effects of corticosterone on 5-HT1A receptor-mediated autoinhibition in the rat dorsal raphe nucleus. Neuropharmacology. 2003;45:925–34. doi: 10.1016/s0028-3908(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, et al. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–8. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Guan Z, Yang X, et al. Impairments of ERK signal transduction in the brain in a rat model of depression induced by neonatal exposure of clomipramine. Brain Res. 2003;991:195–205. doi: 10.1016/j.brainres.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Galeotti N, Bartolini A, Ghelardini C. Role of Gi proteins in the antidepressant-like effect of amitriptyline and clomipramine. Neuropsychopharmacology. 2002;27:554–64. doi: 10.1016/S0893-133X(02)00340-8. [DOI] [PubMed] [Google Scholar]
- Galter D, Unsicker K. Brain-derived neurotrophic factor and trkB are essential for cAMP-mediated induction of the serotonergic neuronal phenotype. J Neurosci Res. 2000;61:295–301. doi: 10.1002/1097-4547(20000801)61:3<295::AID-JNR7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Gill CH, Soffin EM, Hagan JJ, et al. 5-HT7 receptors modulate synchronized network activity in rat hippocampus. Neuropharmacology. 2002;42:82–92. doi: 10.1016/s0028-3908(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Gilmor ML, Owens MJ, Nemeroff CB. Inhibition of norepinephrine uptake in patients with major depression treated with paroxetine. Am J Psychiatry. 2002;159:1702–10. doi: 10.1176/appi.ajp.159.10.1702. [DOI] [PubMed] [Google Scholar]
- Gloyn AL, Desa IM, Clark A, et al. Human calcium/calmodulin-dependent protein kinase II gamma gene (CAMK2G): cloning, genomic structure and detection of variants in subjects with type II diabetes. Diabetologia. 2002;45:580–3. doi: 10.1007/s00125-002-0779-8. [DOI] [PubMed] [Google Scholar]
- Gold PW, Chrousos GP. Organization of the stress system and its dysregulation in melancholic and atypical depression: high vs low CRH/NE states. Mol Psychiatry. 2002;7:254–75. doi: 10.1038/sj.mp.4001032. [DOI] [PubMed] [Google Scholar]
- Gold PW, Goodwin FK, Chrousos GP. Clinical and biochemical manifestations of depression: relation to the neurobiology of stress (1) N Engl J Med. 1988;319:348–53. doi: 10.1056/NEJM198808113190606. [DOI] [PubMed] [Google Scholar]
- Gomez LL, Alam S, Smith KE, et al. Regulation of A-kinase anchoring protein 79/150-cAMP-dependent protein kinase postsynaptic targeting by NMDA receptor activation of calcineurin and remodeling of dendritic actin. J Neurosci. 2002;22:7027–44. doi: 10.1523/JNEUROSCI.22-16-07027.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gay MA, Hajeer AH, Garcia-Porrua C, et al. Corticotropin-releasing hormone promoter polymorphisms in patients with rheumatoid arthritis from northwest Spain. J Rheumatol. 2003;30:913–17. [PubMed] [Google Scholar]
- Gould TD, Manji HK. Signaling networks in the pathophysiology and treatment of mood disorders. J Psychosom Res. 2002;53:687–97. doi: 10.1016/s0022-3999(02)00426-9. [DOI] [PubMed] [Google Scholar]
- Gurguis GN, Vo SP, Griffith JM, et al. Platelet alpha2A-adrenoceptor function in major depression: Gi coupling, effects of imipramine and relationship to treatment outcome. Psychiatry Res. 1999;89:73–95. doi: 10.1016/s0165-1781(99)00103-1. [DOI] [PubMed] [Google Scholar]
- Hawi Z, Dring M, Kirley A, et al. Serotonergic system and attention deficit hyperactivity disorder (ADHD): a potential susceptibility locus at the 5-HT(1B) receptor gene in 273 nuclear families from a multi-centre sample. Mol Psychiatry. 2002;7:718–25. doi: 10.1038/sj.mp.4001048. [DOI] [PubMed] [Google Scholar]
- Heils A, Mossner R, Lesch KP. The human serotonin transporter gene polymorphism – basic research and clinical implication. J Neural Transm. 1997;104:1005–14. doi: 10.1007/BF01273314. [DOI] [PubMed] [Google Scholar]
- Hellevuo K, Welborn R, Menninger JA, et al. Human adenylyl cyclase type 7 contains polymorphic repeats in the 3′ untranslated region: investigations of association with alcoholism. Am J Med Genet. 1997;74:95–8. doi: 10.1002/(sici)1096-8628(19970221)74:1<95::aid-ajmg19>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Helmeste DM, Tang SW. Tyrosine kinase inhibitors regulate serotonin uptake in platelets. Eur J Pharmacol. 1995;280:R5–7. doi: 10.1016/0014-2999(95)00323-d. [DOI] [PubMed] [Google Scholar]
- Herr AS, Tsolakidou AF, Yassouridis A, et al. Antidepressants differentially influence the transcriptional activity of the glucocorticoid receptor in vitro. Neuroendocrinology. 2003;78:12–22. doi: 10.1159/000071701. [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Timmermann B, Lehrach H. Human inter-individual DNA sequence variation in candidate genes, drug targets, the importance of haplotypes and pharmacogenomics. Curr Pharm Biotechnol. 2003;4:351–78. doi: 10.2174/1389201033377300. [DOI] [PubMed] [Google Scholar]
- Holmes C, Arranz M, Collier D, et al. Depression in Alzheimer’s disease: the effect of serotonin receptor gene variation. Am J Med Genet. 2003;119B:40–3. doi: 10.1002/ajmg.b.10068. [DOI] [PubMed] [Google Scholar]
- Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology. 2000;23:477–501. doi: 10.1016/S0893-133X(00)00159-7. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Huo SJ, Yen FC, et al. Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003;48:186–9. doi: 10.1159/000074636. [DOI] [PubMed] [Google Scholar]
- Huizenga NATM, Koper JW, De Lange P, et al. A polymorphism in the glucocorticoid receptor gene may be associated with an increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab. 1998;83:144–51. doi: 10.1210/jcem.83.1.4490. [DOI] [PubMed] [Google Scholar]
- Ito K, Yoshida K, Sato K, et al. A variable number of tandem repeats in the serotonin transporter gene does not affect the antidepressant response to fluvoxamine. Psychiatry Res. 2002;111:235–9. doi: 10.1016/s0165-1781(02)00141-5. [DOI] [PubMed] [Google Scholar]
- Iversen L. Neurotransmitter transporters: fruitful targets for CNS drug discovery. Mol Psychiatry. 2000;5:357–62. doi: 10.1038/sj.mp.4000728. [DOI] [PubMed] [Google Scholar]
- Jensen JB, Mikkelsen JD, Mork A. Increased adenylyl cyclase type 1 mRNA, but not adenylyl cyclase type 2 in the rat hippocampus following antidepressant treatment. Eur Neuropsychopharmacol. 2000;10:105–11. doi: 10.1016/s0924-977x(99)00064-4. [DOI] [PubMed] [Google Scholar]
- Jia H, Hingorani AD, Sharma P, et al. Association of the G(s)alpha gene with essential hypertension and response to beta-blockade. Hypertension. 1999;34:8–14. doi: 10.1161/01.hyp.34.1.8. [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Cichon S, Gustavsson JP, et al. Association between a promoter dopamine D2 receptor gene variant and the personality trait detachment. Biol Psychiatry. 2003;53:577–84. doi: 10.1016/s0006-3223(02)01732-8. [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Nothen MM, Gustavsson JP, et al. Polymorphisms in the dopamine, serotonin, and norepinephrine transporter genes and their relationships to monoamine metabolite concentrations in CSF of healthy volunteers. Psychiatry Res. 1998;79:1–9. doi: 10.1016/s0165-1781(98)00027-4. [DOI] [PubMed] [Google Scholar]
- Judson R. Using multiple drug exposure levels to optimize power in pharmacogenetic trials. J Clin Pharmacol. 2003;43:816–24. doi: 10.1177/0091270003254801. [DOI] [PubMed] [Google Scholar]
- Keck ME, Welt T, Muller MB, et al. Reduction of hypothalamic vasopressinergic hyperdrive contributes to clinically relevant behavioral and neuroendocrine effects of chronic paroxetine treatment in a psychopathological rat model. Neuropsychopharmacology. 2003;28:235–43. doi: 10.1038/sj.npp.1300040. [DOI] [PubMed] [Google Scholar]
- Kendell RE. Five criteria for an improved taxonomy of mental disorders. In: Helzer JE, Hudziak JJ, editors. Defining psychopathology in the 21st Century. Washington: American Psychiatric Publ; 2001. pp. 3–18. [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, et al. A population-based twin study of major depression in women. The impact of varying definitions of illness. Arch Gen Psychiatry. 1992;49:257–66. doi: 10.1001/archpsyc.1992.01820040009001. [DOI] [PubMed] [Google Scholar]
- Kennedy MB. Signal transduction molecules at the glutamatergic postsynaptic membrane. Brain Res Brain Res Rev. 1998;26:243–57. doi: 10.1016/s0165-0173(97)00043-x. [DOI] [PubMed] [Google Scholar]
- Kim DK, Lim S-W, Lee S, et al. Serotonin transporter gene polymorphism and antidepressant response. Neuroreport. 2000;11:215–19. doi: 10.1097/00001756-200001170-00042. [DOI] [PubMed] [Google Scholar]
- Klee CB, Ren H, Wang X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem. 1998;273:13367–70. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- Koch JM, Kell S, Hinze-Selch D, et al. Changes in CREB-phosphorylation during recovery from major depression. J Psychiatr Res. 2002;36:369–75. doi: 10.1016/s0022-3956(02)00056-0. [DOI] [PubMed] [Google Scholar]
- Kohen R, Neumaier J, Hamblin M, et al. Congenitally learned helpless rats show abnormalities in intracellular signaling. Biol Psychiatry. 2003;53:520–9. doi: 10.1016/s0006-3223(02)01503-2. [DOI] [PubMed] [Google Scholar]
- Kopnisky KL, Cowan WM, Hyman SE. Levels of analysis in psychiatric research. Dev Psychopathol. 2002;14:437–61. doi: 10.1017/s0954579402003036. [DOI] [PubMed] [Google Scholar]
- Kunzel HE, Zobel AW, Nickel T, et al. Treatment of depression with the CRH-1-receptor antagonist R121919: endocrine changes and side effects. J Psychiatr Res. 2003;37:525–33. doi: 10.1016/s0022-3956(03)00070-0. [DOI] [PubMed] [Google Scholar]
- Landgrebe J, Welzl G, Metz T, et al. Molecular characterisation of antidepressant effects in the mouse brain using gene expression profiling. J Psychiatr Res. 2002;36:119–29. doi: 10.1016/s0022-3956(01)00061-9. [DOI] [PubMed] [Google Scholar]
- Laplante P, Diorio J, Meaney MJ. Serotonin regulates hippocampal glucocorticoid receptor expression via a 5-HT7 receptor. Brain Res Dev Brain Res. 2002;139:199–203. doi: 10.1016/s0165-3806(02)00550-3. [DOI] [PubMed] [Google Scholar]
- Lemonde S, Turecki G, Bakish D, et al. Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci. 2003;23:8788–99. doi: 10.1523/JNEUROSCI.23-25-08788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard BE. New approaches to the treatment of depression. J Clin Psychiatry. 1996;57:26–33. [PubMed] [Google Scholar]
- Lesch KP. Serotonin transporter and psychiatric disorders: listening to the gene. Neuroscientist. 1998;4:25–34. [Google Scholar]
- Lesch KP, Hough CJ, Aulakh CS, et al. Fluoxetine modulates G protein alpha s, alpha q, alpha 12 subunit mRNA expression in rat brain. Eur J Pharmacol. 1992;227:233–7. doi: 10.1016/0922-4106(92)90134-h. [DOI] [PubMed] [Google Scholar]
- Levitan IB. Modulation of ion channels by protein phosphorylation and dephosphorylation. Annu Rev Physiol. 1994;56:193–212. doi: 10.1146/annurev.ph.56.030194.001205. [DOI] [PubMed] [Google Scholar]
- Li X, Witkin JM, Need AB, et al. Enhancement of antidepressant potency by a potentiator of AMPA receptors. Cell Mol Neurobiol. 2003;23:419–30. doi: 10.1023/A:1023648923447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotrich FE, Pollock BG. Meta-analysis of serotonin transporter polymorphisms and affective disorder. Psychiatr Genet. 2004 doi: 10.1097/00041444-200409000-00001. In press. [DOI] [PubMed] [Google Scholar]
- Lotrich FE, Pollock BG, Ferrell RE. Polymorphism of the serotonin transporter: implications for the use of selective serotonin reuptake inhibitors. Am J Pharmacogenomics. 2001;1:153–64. doi: 10.2165/00129785-200101030-00001. [DOI] [PubMed] [Google Scholar]
- Lotrich FE, Pollock BG, Ferrell RE. Serotonin transporter promoter polymorphism in African Americans. Am J Pharmacogenomics. 2003;3:145–7. doi: 10.2165/00129785-200303020-00007. [DOI] [PubMed] [Google Scholar]
- Lovlie R, Berle JO, Stordal E, et al. The phospholipase C-gamma1 gene (PLCG1) and lithium-responsive bipolar disorder: re-examination of an intronic dinucleotide repeat polymorphism. Psychiatr Genet. 2001;11:41–3. doi: 10.1097/00041444-200103000-00008. [DOI] [PubMed] [Google Scholar]
- Lucek P, Hanke J, Reich J, et al. Multilocus nonparametric linkage analysis of complex trait loci with neural network. Hum Hered. 1998;48:275–84. doi: 10.1159/000022816. [DOI] [PubMed] [Google Scholar]
- Malberg JE, Eisch AJ, Nestler EJ, et al. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci. 2000;20:9104–10. doi: 10.1523/JNEUROSCI.20-24-09104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancama D, Kerwin RW. Role of pharmacogenomics in individualising treatment with SSRIs. CNS Drugs. 2003;17:143–51. doi: 10.2165/00023210-200317030-00001. [DOI] [PubMed] [Google Scholar]
- Manji HK, Chen G. PKC, MAP kinases and the bcl-2 family of proteins as long-term targets for mood stabilizers. Mol Psychiatry. 2002;7:S46–56. doi: 10.1038/sj.mp.4001018. [DOI] [PubMed] [Google Scholar]
- Manji HK, Drevets WC, Charney DS. The cellular neurobiology of depression. Nat Med. 2001;7:541–7. doi: 10.1038/87865. [DOI] [PubMed] [Google Scholar]
- Mann CD, Vu TB, Hrdina PD. Protein kinase C in rat brain cortex and hippocampus: effect of repeated administration of fluoxetine and desipramine. Br J Pharmacol. 1995;115:595–600. doi: 10.1111/j.1476-5381.1995.tb14973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek GJ, Carpenter LL, McDougle CJ, et al. Synergistic action of 5-HT2A antagonists and selective serotonin reuptake inhibitors in neuropsychiatric disorders. Neuropsychopharmacology. 2003;28:402–12. doi: 10.1038/sj.npp.1300057. [DOI] [PubMed] [Google Scholar]
- Mason DA, Moore JD, Green SA, et al. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem. 1999;274:12670–4. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- Massat I, Souery D, Del-Favero J, et al. Positive association of dopamine D2 receptor polymorphism with bipolar affective disorder in a European Multicenter Association Study of affective disorders. Am J Med Genet. 2002;114:177–85. [PubMed] [Google Scholar]
- McCarthy JJ, Hilfiker R. The use of single-nucleotide polymorphism maps in pharmacogenomics. Nat Biotechnol. 2000;18:505–8. doi: 10.1038/75360. [DOI] [PubMed] [Google Scholar]
- McLeod TM, Lopez-Figueroa AL, Lopez-Figueroa MO. Nitric oxide, stress, and depression. Psychopharmacol Bull. 2001;35:24–41. [PubMed] [Google Scholar]
- Meira-Lima I, Jardim D, Junqueira R, et al. Allelic association study between phospholipase A2 genes and bipolar affective disorder. Bipolar Disord. 2003;5:295–9. doi: 10.1034/j.1399-5618.2003.00025.x. [DOI] [PubMed] [Google Scholar]
- Messa C, Colombo C, Moresco RM, et al. 5-HT(2A) receptor binding is reduced in drug-naive and unchanged in SSRI-responder depressed patients compared to healthy controls: a PET study. Psychopharmacology (Berl) 2003;167:72–8. doi: 10.1007/s00213-002-1379-5. [DOI] [PubMed] [Google Scholar]
- Messina D, Annesi G, Serra P, et al. Association of the 5-HT6 receptor gene polymorphism C267T with Parkinson’s disease. Neurology. 2002;58:828–9. doi: 10.1212/wnl.58.5.828. [DOI] [PubMed] [Google Scholar]
- Michaelovsky E, Frisch A, Rockah R, et al. A novel allele in the promoter region of the human serotonin transporter gene. Mol Psychiatry. 1999;4:97–9. doi: 10.1038/sj.mp.4000449. [DOI] [PubMed] [Google Scholar]
- Millan MJ, Veiga S, Girardon S, et al. Blockade of serotonin 5-HT1B and 5-HT2A receptors suppresses the induction of locomotor activity by 5-HT reuptake inhibitors, citalopram and fluvoxamine, in NMRI mice exposed to a novel environment: a comparison to other 5-HT receptor subtypes. Psychopharmacology (Berl) 2003;168:397–409. doi: 10.1007/s00213-003-1389-y. [DOI] [PubMed] [Google Scholar]
- Minov C, Baghai TC, Schule C, et al. Serotonin-2A-receptor and -transporter polymorphisms: lack of association in patients with major depression. Neurosci Lett. 2001;303:119–22. doi: 10.1016/s0304-3940(01)01704-9. [DOI] [PubMed] [Google Scholar]
- Miro X, Perez-Torres S, Artigas F, et al. Regulation of cAMP phosphodiesterase mRNAs expression in rat brain by acute and chronic fluoxetine treatment. An in situ hybridization study. Neuropharmacology. 2002;43:1148–57. doi: 10.1016/s0028-3908(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Mitchell RJ, Howlett S, Earl L, et al. Distribution of the 3′ VNTR polymorphism in the human dopamine transporter gene in world populations. Hum Biol. 2000;72:295–304. [PubMed] [Google Scholar]
- Miyamoto S, Asakura M, Sasuga Y, et al. Effects of long-term treatment with desipramine on microtubule proteins in rat cerebral cortex. Eur J Pharmacol. 1997;333:279–87. doi: 10.1016/s0014-2999(97)01140-0. [DOI] [PubMed] [Google Scholar]
- Monaca C, Boutrel B, Hen R, et al. 5-HT1a/1b receptor-mediated effects of the selective serotonin reuptake inhibitor, citalopram, on sleep: studies in 5-HT1a and 5-HT1b knockout mice. Neuropsychopharmacology. 2003;28:850–6. doi: 10.1038/sj.npp.1300109. [DOI] [PubMed] [Google Scholar]
- Moretti A, Gorini A, Villa RF. Affective disorders, antidepressant drugs and brain metabolism. Mol Psychiatry. 2003;8:773–85. doi: 10.1038/sj.mp.4001353. [DOI] [PubMed] [Google Scholar]
- Mori S, Garbini S, Caivano M, et al. Time-course changes in rat cerebral cortex subcellular distribution of the cyclic-AMP binding after treatment with selective serotonin reuptake inhibitors. Int J Neuropsychopharmacol. 1998;1:3–10. doi: 10.1017/S1461145798001084. [DOI] [PubMed] [Google Scholar]
- Mossner R, Daniel S, Albert D, et al. Serotonin transporter function is modulated by brain-derived neurotrophic factor (BDNF) but not nerve growth factor (NGF) Neurochem Int. 2000;36:197–202. doi: 10.1016/s0197-0186(99)00122-9. [DOI] [PubMed] [Google Scholar]
- Mundo E, Tharmalingham S, Neves-Pereira M, et al. Evidence that the N-methyl-D-aspartate subunit 1 receptor gene (GRIN1) confers susceptibility to bipolar disorder. Mol Psychiatry. 2003;8:241–5. doi: 10.1038/sj.mp.4001218. [DOI] [PubMed] [Google Scholar]
- Mundo E, Walker M, Cate T, et al. The role of serotonin transporter protein gene in antidepressant-induced mania in bipolar disorder: preliminary findings. Arch Gen Psychiatry. 2001;58:539–44. doi: 10.1001/archpsyc.58.6.539. [DOI] [PubMed] [Google Scholar]
- Murphy DL, Uhl GR, Holmes A, et al. Experimental gene interaction studies with SERT mutant mice as models for human polygenic and epistatic traits and disorders. Genes Brain Behav. 2003;2:359–64. doi: 10.1046/j.1601-1848.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- Murphy GM, Kremer C, Rodrigues H, et al. The apolipoprotein E epsilon4 allele and antidepressant efficacy in cognitively intact elderly depressed patients. Biol Psychiatry. 2003a;54:665–73. doi: 10.1016/s0006-3223(03)00174-4. [DOI] [PubMed] [Google Scholar]
- Murphy GM, Jr, Kremer C, Rodrigues HE, et al. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry. 2003b;160:1830–5. doi: 10.1176/appi.ajp.160.10.1830. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ueno S, Sano A, et al. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol Psychiatry. 2000;5:32–8. doi: 10.1038/sj.mp.4000698. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Terwilliger RZ, Duman RS. Chronic antidepressant administration alters the subcellular distribution of cyclic AMP-dependent protein kinase in rat frontal cortex. J Neurochem. 1989;53:1644–7. doi: 10.1111/j.1471-4159.1989.tb08564.x. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Mundo E, Muglia P, et al. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002;71:651–5. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton SS, Thome J, Wallace TL, et al. Inhibition of cAMP response element-binding protein or dynorphin in the nucleus accumbens produces an antidepressant-like effect. J Neurosci. 2002;24:10883–90. doi: 10.1523/JNEUROSCI.22-24-10883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Trakalo JM, Mundo E, et al. Linkage disequilibrium between dopamine D1 receptor gene (DRD1) and bipolar disorder. Biol Psychiatry. 2002;52:1144–50. doi: 10.1016/s0006-3223(02)01433-6. [DOI] [PubMed] [Google Scholar]
- Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15:7539–47. doi: 10.1523/JNEUROSCI.15-11-07539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibuya M, Nestler EJ, Duman RS. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci. 1996;16:2365–72. doi: 10.1523/JNEUROSCI.16-07-02365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierenberg AA. Predictors of response to antidepressants general principles and clinical implications. Psychiatr Clin North Am. 2003;26:345–52. doi: 10.1016/s0193-953x(02)00105-3. [DOI] [PubMed] [Google Scholar]
- Nierenberg AA, DeCecco LM. Definitions of antidepressant treatment response, remission, nonresponse, partial response, and other relevant outcomes: a focus on treatment-resistant depression. J Clin Psychiatry. 2001;62:5–9. [PubMed] [Google Scholar]
- Niesler B, Flohr T, Nothen MM, et al. Association between the 5′ UTR variant C178T of the serotonin receptor gene HTR3A and bipolar affective disorder. Pharmacogenetics. 2001;11:471–5. doi: 10.1097/00008571-200108000-00002. [DOI] [PubMed] [Google Scholar]
- Nunoki K, Florio V, Catterall WA. Activation of purified calcium channels by stoichiometric protein phosphorylation. Proc Natl Acad Sci U S A. 1989;86:6818–20. doi: 10.1073/pnas.86.17.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger J, Dammer S, Mitchell A, et al. Effect of the C825T polymorphism of the G protein beta 3 subunit on the systolic blood pressure-lowering effect of clonidine in young, healthy male subjects. Clin Pharmacol Therap. 2003;74:53–60. doi: 10.1016/S0009-9236(03)00087-0. [DOI] [PubMed] [Google Scholar]
- Odagaki Y, Garcia-Sevilla JA, Huguelet P, et al. Cyclic AMP-mediated signaling components are upregulated in the prefrontal cortex of depressed suicide victims. Brain Res. 2001;898:224–31. doi: 10.1016/s0006-8993(01)02188-6. [DOI] [PubMed] [Google Scholar]
- Ordway GA, Klimek V, Mann JJ. Neurocircuitry of mood disorders. In: Davis KL, Charney D, Coyle JT, et al., editors. Neuropsychopharmacology: the fifth generation of progress. Philadelphia: Lippincott Williams & Williams; 2002. pp. 1051–64. [Google Scholar]
- O’Reilly RL, Bogue L, Singh SM. Pharmacogenetic response to antidepressants in a multicase family with affective disorder. Biol Psychiatry. 1994;36:467–71. doi: 10.1016/0006-3223(94)90642-4. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Carr LG, Heilig M, et al. Neuropeptide Y and alcoholism: genetic, molecular, and pharmacological evidence. Alcohol Clin Exp Res. 2003;27:149–54. doi: 10.1097/01.ALC.0000052706.21367.0E. [DOI] [PubMed] [Google Scholar]
- Papadimitriou GN, Dikeos DG, Souery D, et al. Genetic association between the phospholipase A2 gene and unipolar affective disorder: a multicentre case-control study. Psychiatr Genet. 2003;13:211–20. doi: 10.1097/00041444-200312000-00004. [DOI] [PubMed] [Google Scholar]
- Pare CM, Mack JW. Differentiation of two genetically specific types of depression by the response to antidepressant drugs. J Med Genet. 1971;8:306–9. doi: 10.1136/jmg.8.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G, Anderson IM, Haddad P. Clinical trials of antidepressant medications are producing meaningless results. Br J Psychiatry. 2002;183:102–4. doi: 10.1192/bjp.183.2.102. [DOI] [PubMed] [Google Scholar]
- Patel PD, Pontrello C, Burke S. Robust and tissue-specific expression of TPH2 versus TPH1 in rat raphe and pineal gland. Biol Psychiatry. 2004;55:428–33. doi: 10.1016/j.biopsych.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Perez J, Tinelli D, Bianchi E, et al. cAMP binding proteins in the rat cerebral cortex after administration of selective 5-HT and NE reuptake blockers with antidepressant activity. Neuropsychopharmacology. 1991;4:57–64. [PubMed] [Google Scholar]
- Phillips TJ. Behavior genetics of drug sensitization. Crit Rev Neurobiol. 1997;11:21–33. doi: 10.1615/critrevneurobiol.v11.i1.20. [DOI] [PubMed] [Google Scholar]
- Pickar D, Rubinow K. Pharmacogenomics of psychiatric disorders. Trends Pharmacol Sci. 2001;22:75–83. doi: 10.1016/s0165-6147(00)01603-5. [DOI] [PubMed] [Google Scholar]
- Pilc A, Branski P, Palucha A, et al. The effect of prolonged imipramine and electroconvulsive shock treatment on calcium/calmodulin-dependent protein kinase II in the hippocampus of rat brain. Neuropharmacology. 1999;38:597–603. doi: 10.1016/s0028-3908(98)00211-1. [DOI] [PubMed] [Google Scholar]
- Pliakas A, Carlson RR, Neve RL, et al. Altered responsiveness to cocaine and increased immobility in the forced swim test associated with elevated CREB expression in the nucleus accumbens. J Neurosci. 2001;21:7397–403. doi: 10.1523/JNEUROSCI.21-18-07397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock BG, Ferrell RE, Mulsant BH, et al. Allelic variation in the serotonin transporter promoter affects onset of paroxetine treatment response in late-life depression. Neuropsychopharmacology. 2000;23:587–90. doi: 10.1016/S0893-133X(00)00132-9. [DOI] [PubMed] [Google Scholar]
- Popoli M, Vocaturo C, Perez J, et al. Presynaptic Ca2+/calmodulin-dependent protein kinase II: autophosphorylation and activity increase in the hippocampus after long-term blockade of serotonin reuptake. Mol Pharmacol. 1995;48:623–9. [PubMed] [Google Scholar]
- Price NE, Mumbly MC. Brain protein serine/threonine phosphatases. Curr Opin Neurobiol. 1999;9:336–42. doi: 10.1016/s0959-4388(99)80049-x. [DOI] [PubMed] [Google Scholar]
- Qian Y, Galli A, Ramamoorthy S, et al. Protein kinase C activation regulates human serotonin transporters in HEK-293 cells via altered cell surface expression. J Neurosci. 1997;17:45–57. doi: 10.1523/JNEUROSCI.17-01-00045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Chang L, Klaff J, et al. Imaging of brain serotonergic neurotransmission involving phospholipase A2 activation and arachidonic acid release in unanesthetized rats. Brain Res Brain Res Protoc. 2003a;12:16–25. doi: 10.1016/s1385-299x(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Qu Y, Chang L, Klaff J, et al. Imaging brain phospholipase A2-mediated signal transduction in response to acute fluoxetine administration in unanesthetized rats. Neuropsychopharmacology. 2003b;28:1219–26. doi: 10.1038/sj.npp.1300177. [DOI] [PubMed] [Google Scholar]
- Rahimian R, Hrdina PD. Possible role of protein kinase C in regulation of 5-hydroxytryptamine 2A receptors in rat brain. Can J Physiol Pharmacol. 1995;73:1686–91. doi: 10.1139/y95-731. [DOI] [PubMed] [Google Scholar]
- Rahman S, Li PP, Young LT, et al. Reduced [3H]cyclic AMP binding in postmortem brain from subjects with bipolar affective disorder. J Neurochem. 1997;68:297–304. doi: 10.1046/j.1471-4159.1997.68010297.x. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S, Giovanetti E, Qian Y, et al. Phosphorylation and regulation of antidepressant-sensitive serotonin transporters. J Biol Chem. 1998;273:2458–66. doi: 10.1074/jbc.273.4.2458. [DOI] [PubMed] [Google Scholar]
- Ranade K, Jorgenson E, Sheu WH, et al. A polymorphism in the beta1 adrenergic receptor is associated with resting heart rate. Am J Hum Genet. 2002;70:935–42. doi: 10.1086/339621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasenick MM, Chaney KA, Chen J. G protein-mediated signal transduction as a target of antidepressant and antibipolar drug action: evidence from model systems. J Clin Psychiatry. 1996;57:49–55. [PubMed] [Google Scholar]
- Rausch JL, Gillespie CF, Fei Y, et al. Antidepressant effect on kinase gene expression patterns in rat brain. Neurosci Lett. 2002;334:91–4. doi: 10.1016/s0304-3940(02)01106-0. [DOI] [PubMed] [Google Scholar]
- Rausch JL, Johnson ME, Fei Y-J, et al. Initial conditions of serotonin transporter kinetics and genotype: influence on SSRI treatment trial outcome. Biol Psychiatry. 2002;51:723–32. doi: 10.1016/s0006-3223(01)01283-5. [DOI] [PubMed] [Google Scholar]
- Raymond JR, Mukhin YV, Gelasco A, et al. Multiplicity of mechanisms of serotonin receptor signal transduction. Pharmacol Therap. 2001;92:179–212. doi: 10.1016/s0163-7258(01)00169-3. [DOI] [PubMed] [Google Scholar]
- Redrobe JP, Bourin M. Partial role of 5-HT2 and 5-HT3 receptors in the activity of antidepressants in the mouse forced swimming test. Eur J Pharmacol. 1997;325:129–35. doi: 10.1016/s0014-2999(97)00115-5. [DOI] [PubMed] [Google Scholar]
- Riccio A, Ahn S, Davenport CM, et al. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999;286:2358–61. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- Richards JG, Saura J, Luque JM, et al. Monoamine oxidases: from brain maps to physiology and transgenics to pathophysiology. J Neural Transm. 1998;52(Suppl):173–87. doi: 10.1007/978-3-7091-6499-0_17. [DOI] [PubMed] [Google Scholar]
- Robinson-White A, Stratakis C. Protein kinase A signaling: “crosstalk” with other pathways in endocrine cells. Ann N Y Acad Sci. 2002;968:256–70. doi: 10.1111/j.1749-6632.2002.tb04340.x. [DOI] [PubMed] [Google Scholar]
- Rogoz Z, Skuza G, Maj J, et al. Synergistic effect of uncompetitive NMDA receptor antagonists and antidepressant drugs in the forced swimming test in rats. Neuropharmacology. 2002;42:1024–30. doi: 10.1016/s0028-3908(02)00055-2. [DOI] [PubMed] [Google Scholar]
- Rosin DL, Melia K, Knorr AM, et al. Chronic imipramine administration alters the activity and phosphorylation state of tyrosine hydroxylase in dopaminergic regions of rat brain. Neuropsychopharmacology. 1995;12:113–21. doi: 10.1016/0893-133X(94)00066-9. [DOI] [PubMed] [Google Scholar]
- Rosmond R, Bouchard C, Bjorntorp P. A C-1291G polymorphism in the alpha2A-adrenergic receptor gene (ADRA2A) promoter is associated with cortisol escape from dexamethasone and elevated glucose levels. J Intern Med. 2002;251:252–7. doi: 10.1046/j.1365-2796.2002.00961.x. [DOI] [PubMed] [Google Scholar]
- Rosmond R, Chagnon M, Bouchard C, et al. A polymorphism in the regulatory region of the corticotropin-releasing hormone gene in relation to cortisol secretion, obesity, and gene-gene interaction. Metab Clin Exp. 2001;50:1059–62. doi: 10.1053/meta.2001.25598. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Giegling I, Sato T, et al. Genetic variations in tryptophan hydroxylase in suicidal behavior: analysis and meta-analysis. Biol Psychiatry. 2003;54:465–73. doi: 10.1016/s0006-3223(02)01748-1. [DOI] [PubMed] [Google Scholar]
- Rumajogee P, Madeira A, Verge D, et al. Up-regulation of the neuronal serotoninergic phenotype in vitro: BDNF and cAMP share Trk B-dependent mechanisms. J Neurochem. 2002;83:1525–8. doi: 10.1046/j.1471-4159.2002.01264.x. [DOI] [PubMed] [Google Scholar]
- Saarelainen T, Hendolin P, Lucas G, et al. Activation of the trkB neurotrophin receptor is induced by antidepressant drugs and is required for antidepressant-induced behavioral effects. J Neurosci. 2003;23:349–57. doi: 10.1523/JNEUROSCI.23-01-00349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–9. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Saito N, Shirai Y. Protein kinase C gamma (PKC gamma): function of neuron specific isotype. J Biochem. 2002;132:683–7. doi: 10.1093/oxfordjournals.jbchem.a003274. [DOI] [PubMed] [Google Scholar]
- Sanacora G, Rothman DL, Mason G, et al. Clinical studies implementing glutamate neurotransmission in mood disorders. Ann N Y Acad Sci. 2003;1003:292–308. doi: 10.1196/annals.1300.018. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Gejman PV. DNA variation and psychopharmacology of the human serotonin receptor 1B (HTR1B) gene. Pharmacogenomics. 2002;3:745–62. doi: 10.1517/14622416.3.6.745. [DOI] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–9. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Sato K, Yoshida K, Takahashi H, et al. Association between -1438G/A promoter polymorphism in the 5-HT(2A) receptor gene and fluvoxamine response in Japanese patients with major depressive disorder. Neuropsychobiology. 2002;46:136–40. doi: 10.1159/000066394. [DOI] [PubMed] [Google Scholar]
- Saura J, Bleuel Z, Ulrich J, et al. Molecular neuroanatomy of human monoamine oxidases A and B revealed by quantitative enzyme radioautography and in situ hybridization histochemistry. Neuroscience. 1996;70:755–74. doi: 10.1016/s0306-4522(96)83013-2. [DOI] [PubMed] [Google Scholar]
- Schaefer ML, Wong ST, Wozniak DF, et al. Altered stress-induced anxiety in adenylyl cyclase type VIII-deficient mice. J Neurosci. 2000;20:4809–20. doi: 10.1523/JNEUROSCI.20-13-04809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzberg AF, Garlow SJ, Nemeroff CB. Molecular and cellular mechanisms in depression. In: Davis KL, Charney D, Coyle JT, et al., editors. Neuropsychopharmacology: the fifth generation of progress. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 1039–50. [Google Scholar]
- Schulze TG, Hardy J, McMahon FJ. Inconsistent designs of association studies: a missed opportunity. Mol Psychiatry. 2003;8:770–2. doi: 10.1038/sj.mp.4001329. [DOI] [PubMed] [Google Scholar]
- Scott JD. Dissection of protein kinase and phosphatase targeting interactions. Soc Gen Physiol Ser. 1997;52:227–39. [PubMed] [Google Scholar]
- Selinger-Leneman H, Genin E, Norris JM, et al. Does accounting for gene-environment (GxE) interaction increase the power to detect the effect of a gene in a multifactorial disease? Genet Epidemiol. 2003;24:200–7. doi: 10.1002/gepi.10221. [DOI] [PubMed] [Google Scholar]
- Serretti A, Franchini L, Gasperini M, et al. Mode of inheritance in mood disorders families according to fluvoxamine response. Acta Psychiatr Scand. 1998;98:443–50. doi: 10.1111/j.1600-0447.1998.tb10117.x. [DOI] [PubMed] [Google Scholar]
- Serretti A, Lilli R, Smeraldi E. Pharmacogenetics in affective disorders. Eur J Pharmacol. 2002;438:117–28. doi: 10.1016/s0014-2999(02)01309-2. [DOI] [PubMed] [Google Scholar]
- Serretti A, Lorenz C, Cusin C, et al. SSRIs antidepressant activity is influenced by G beta 3 variants. Eur Neuropsychopharmacol. 2003;13:117–22. doi: 10.1016/s0924-977x(02)00154-2. [DOI] [PubMed] [Google Scholar]
- Serretti A, Zanardi R, Cusin C, et al. Tryptophan hydroxylase gene associated with paroxetine antidepressant activity. Eur Neuropsychopharmacol. 2001a;11:375–80. doi: 10.1016/s0924-977x(01)00113-4. [DOI] [PubMed] [Google Scholar]
- Serretti A, Zanardi R, Cusin C, et al. No association between dopamine D2 and D4 receptor gene variants and antidepressant activity of two selective serotonin reuptake inhibitors. Psychiatry Res. 2001b;104:195–203. doi: 10.1016/s0165-1781(01)00324-9. [DOI] [PubMed] [Google Scholar]
- Serretti A, Zanardi R, Rossini D, et al. Influence of tryptophan hydroxylase and serotonin transporter genes on fluvoxamine antidepressant activity. Mol Psychiatry. 2001;6:586–92. doi: 10.1038/sj.mp.4000876. [DOI] [PubMed] [Google Scholar]
- Shaffery J, Hoffmann R, Armitage R. The neurobiology of depression: perspectives from animal and human sleep studies. Neuroscientist. 2003;9:82–98. doi: 10.1177/1073858402239594. [DOI] [PubMed] [Google Scholar]
- Sheehan TP, Neve RL, Duman RS, et al. Antidepressant effects of the calcium-activated tyrosine kinase Pyk2 in the lateral septum. Biol Psychiatry. 2003;54:540–51. doi: 10.1016/s0006-3223(02)01815-2. [DOI] [PubMed] [Google Scholar]
- Sheline YI. Neuroimaging studies of mood disorder effects on the brain. Biol Psychiatry. 2003;54:338–52. doi: 10.1016/s0006-3223(03)00347-0. [DOI] [PubMed] [Google Scholar]
- Shelton RC. Intracellular mechanisms of antidepressant drug action. Harv Rev Psychiatry. 2000;8:161–74. [PubMed] [Google Scholar]
- Shelton RC, Manier D, Sulser F. cAMP-dependent protein kinase activity in major depression. Am J Psychiatry. 1996;153:1037–42. doi: 10.1176/ajp.153.8.1037. [DOI] [PubMed] [Google Scholar]
- Shirayama Y, Chen AC-H, Nakagawa S, et al. Brain derived neurotrophic factor produces antidepressant effects in behavioral models of depression. J Neurosci. 2002;22:3251–61. doi: 10.1523/JNEUROSCI.22-08-03251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shytle RD, Silver AA, Lukas RJ, et al. Nicotinic acetylcholine receptors as targets for antidepressants. Mol Psychiatry. 2002;7:525–35. doi: 10.1038/sj.mp.4001035. [DOI] [PubMed] [Google Scholar]
- Sim AT, Scott JD. Targeting of PKA, PKC and protein phosphatases to cellular microdomains. Cell Calcium. 1999;26:209–17. doi: 10.1054/ceca.1999.0072. [DOI] [PubMed] [Google Scholar]
- Skolnick P. Modulation of glutamate receptors: strategies for the development of novel antidepressants. Amino Acids. 2002;23:153–9. doi: 10.1007/s00726-001-0121-7. [DOI] [PubMed] [Google Scholar]
- Smith MA, Kakino S, Altemus M, et al. Stress and antidepressants differentially regulate neurotrophin 3 mRNA expression in the locus coeruleus. Proc Natl Acad Sci U S A. 1995;92:8788–92. doi: 10.1073/pnas.92.19.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Lin A, Bonaccorso S, et al. The inflammatory response system and the availability of plasma tryptophan in patients with primary sleep disorders and major depression. J Affect Disord. 1998;49:211–19. doi: 10.1016/s0165-0327(98)00025-1. [DOI] [PubMed] [Google Scholar]
- Srivastava SK, Nath C. The differential effects of calcium channel blockers in the behavioural despair test in mice. Pharmacol Res. 2000;42:293–7. doi: 10.1006/phrs.2000.0696. [DOI] [PubMed] [Google Scholar]
- Stack S, Zaucha JA, Ebner FF, et al. Brain protein phosphatase 2A: developmental regulation and distinct cellular and subcellular locatization by B subunits. J Comp Neurol. 1998;392:515–27. [PubMed] [Google Scholar]
- Steimer W, Muller B, Leucht S, et al. Pharmacogenetics: a new diagnostic tool in the management of antidepressive drug therapy. Clin Chim Acta. 2001;308:33–41. doi: 10.1016/s0009-8981(01)00423-5. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos SG, Stratakis CA. Human tumors associated with Carney complex and germline PRKAR1A mutations: a protein kinase A disease! FEBS Lett. 2003;546:59–64. doi: 10.1016/s0014-5793(03)00452-6. [DOI] [PubMed] [Google Scholar]
- Strohle A, Holsboer F. Stress responsive neurohormones in depression and anxiety. Pharmacopsychiatry. 2003;36:S207–14. doi: 10.1055/s-2003-45132. [DOI] [PubMed] [Google Scholar]
- Sugie Y, Sugie H, Fukuda T, et al. Studies on the adverse effects of fluvoxamine treatment in children with autistic disorder: correlation with genetic polymorphism in serotonin related genes. [Japanese] No to Hattatsu. 2003;35:233–7. [PubMed] [Google Scholar]
- Suzuki A, Kondo T, Mihara K, et al. The -141C Ins/Del polymorphism in the dopamine D2 receptor gene promoter region is associated with anxiolytic and antidepressive effects during treatment with dopamine antagonists in schizophrenic patients. Pharmacogenetics. 2001;11:545–50. doi: 10.1097/00008571-200108000-00009. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Tzavara ET, Witkin JM, et al. Involvement of striatal and extrastriatal DARPP-32 in biochemical and behavioral effects of fluoxetine (Prozac) Proc Natl Acad Sci U S A. 2002;99:3182–7. doi: 10.1073/pnas.052712799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev. 2002;3:1–7. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- Thase ME. Depression, sleep, and antidepressants. J Clin Psychiatry. 1998;59:55–65. [PubMed] [Google Scholar]
- Thase ME. Effectiveness of antidepressants: comparative remission rates. J Clin Psychiatry. 2003;64:3–7. [PubMed] [Google Scholar]
- Thompson C, Peveler RC, Stephenson D, et al. Compliance with antidepressant medication in the treatment of major depressive disorder in primary care: a randomized comparison of fluoxetine and a tricyclic antidepressant. Am J Psychiatry. 2000;157:338–43. doi: 10.1176/appi.ajp.157.3.338. [DOI] [PubMed] [Google Scholar]
- Torres G, Horowitz JM, Laflamme N, et al. Fluoxetine induces the transcription of genes encoding c-fos, corticotropin-releasing factor and its type 1 receptor in rat brain. Neurosci Lett. 1998;87:463–77. doi: 10.1016/s0306-4522(98)00147-x. [DOI] [PubMed] [Google Scholar]
- Toyota T, Hattori E, Meerabux J, et al. Molecular analysis, mutation screening, and association study of adenylate cyclase type 9 gene (ADCY9) in mood disorders. Am J Med Genet. 2002;114:84–92. doi: 10.1002/ajmg.10117. [DOI] [PubMed] [Google Scholar]
- Turecki G, Grof P, Cavazzoni P, et al. Evidence for a role of phospholipase C-gamma1 in the pathogenesis of bipolar disorder. Mol Psychiatry. 1998;3:534–8. doi: 10.1038/sj.mp.4000447. [DOI] [PubMed] [Google Scholar]
- Ueno S. Genetic polymorphisms of serotonin and dopamine transporters in mental disorders. J Med Investig. 2003;50:25–31. [PubMed] [Google Scholar]
- Urwin RE, Bennetts B, Wilcken B, et al. Anorexia nervosa (restrictive subtype) is associated with a polymorphism in the novel norepinephrine transporter gene promoter polymorphic region. Mol Psychiatry. 2002;7:652–7. doi: 10.1038/sj.mp.4001080. [DOI] [PubMed] [Google Scholar]
- van den Bree MB, Owen MJ. The future of psychiatric genetics. Ann Med. 2003;35:122–34. doi: 10.1080/07853890310010023. [DOI] [PubMed] [Google Scholar]
- van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–42. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vicentic A, Battaglia G, Carroll RI, et al. Serotonin transporter production and degradation rates: studies with RTI-76. Brain Res. 1999;841:1–10. doi: 10.1016/s0006-8993(99)01761-8. [DOI] [PubMed] [Google Scholar]
- Villafuerte SM, Del-Favero J, Adolfsson R, et al. Gene-based SNP genetic association study of the corticotropin-releasing hormone receptor-2 (CRHR2) in major depression. Am J Med Genet. 2002;114:222–6. doi: 10.1002/ajmg.10179. [DOI] [PubMed] [Google Scholar]
- Walsh BT, Seidman SN, Sysko R, et al. Placebo response in studies of major depression: variable, substantial, and growing. JAMA. 2002;287:1840–7. doi: 10.1001/jama.287.14.1840. [DOI] [PubMed] [Google Scholar]
- Wang SJ, Su CF, Kuo YH. Fluoxetine depresses glutamate exocytosis in the rat cerebrocortical nerve terminals (synaptosomes) via inhibition of P/Q-type Ca2+ channels. Synapse. 2003;48:170–7. doi: 10.1002/syn.10200. [DOI] [PubMed] [Google Scholar]
- Way KJ, Chou E, King GL. Identification of PKC-isoform specific biological actions using pharmacological approaches. Trends Pharmacol Sci. 2000;21:181–7. doi: 10.1016/s0165-6147(00)01468-1. [DOI] [PubMed] [Google Scholar]
- Wilde MI, Markham A. Ondansetron. A review of its pharmacology and preliminary clinical findings in novel applications. Drugs. 1996;52:773–94. doi: 10.2165/00003495-199652050-00010. [DOI] [PubMed] [Google Scholar]
- Wischmeyer E, Karschin A. Receptor stimulation causes slow inhibition of IRK1 inwardly rectifying K+ channels by direct protein kinase A-mediated phosphorylation. Proc Natl Acad Sci U S A. 1996;93:5819–23. doi: 10.1073/pnas.93.12.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Huo SJ, Cheng CY, et al. Association study of the 5-HT(6) receptor polymorphism (C267T) and symptomatology and antidepressant response in major depressive disorders. Neuropsychobiology. 2001;44:172–5. doi: 10.1159/000054938. [DOI] [PubMed] [Google Scholar]
- Yamada M, Yamada M, Yamazaki S, et al. Identification of a novel gene with RING-H2 finger motif induced after chronic antidepressant treatment in rat brain. Biochem Biophys Res Commun. 2000;278:150–7. doi: 10.1006/bbrc.2000.3773. [DOI] [PubMed] [Google Scholar]
- Yamamoto KK, Gonzalez GA, Biggs WHI, et al. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–8. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- Ye Y, Jackson K, O’Donnell JM. Effects of repeated antidepressant treatment of type 4A phosphodiesterase (PDE4A) in rat brain. J Neurochem. 2000;74:1257–62. doi: 10.1046/j.1471-4159.2000.741257.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama CT, Sheng Z-H, Catterall WA. Phosphorylation of the synaptic protein interaction site on N-type calcium channels inhibits interactions with SNARE proteins. J Neurosci. 1997;17:6929–38. doi: 10.1523/JNEUROSCI.17-18-06929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ito K, Sato K, et al. Influence of the serotonin transporter gene-linked polymorphic region on the antidepressant response to fluvoxamine in Japanese depressed patients. Prog Neuropsychopharmacol Biol Psychiatry. 2002;26:383–6. doi: 10.1016/s0278-5846(01)00287-1. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Naito S, Takahashi H, et al. Monoamine oxidase: a gene polymorphism, tryptophan hydroxylase gene polymorphism and antidepressant response to fluvoxamine in Japanese patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2002;26:1279–83. doi: 10.1016/s0278-5846(02)00267-1. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Naito S, Takahashi H, et al. Monoamine oxidase A gene polymorphism, 5-HT 2A receptor gene polymorphism and incidence of nausea induced by fluvoxamine. Neuropsychobiology. 2003;48:10–3. doi: 10.1159/000071822. [DOI] [PubMed] [Google Scholar]
- Yoshioka M, Matsumoto M, Togashi H, et al. Central distribution and function of 5-HT6 receptor subtype in the rat brain. Life Sci. 1998;62:1473–7. doi: 10.1016/s0024-3205(98)00092-7. [DOI] [PubMed] [Google Scholar]
- Young EA, Lopez JF, Murphy-Weinberg V, et al. Mineralocorticoid receptor function in major depression. Arch Gen Psychiatry. 2003;60:24–8. doi: 10.1001/archpsyc.60.1.24. [DOI] [PubMed] [Google Scholar]
- Yu YW, Chen TJ, Hong CJ, et al. Association study of the interleukin-1 beta (C-511T) genetic polymorphism with major depressive disorder, associated symptomatology, and antidepressant response. Neuropsychopharmacology. 2003;28:1182–5. doi: 10.1038/sj.npp.1300172. [DOI] [PubMed] [Google Scholar]
- Yu YW, Chen TJ, Wang YC, et al. Association analysis for neuronal nitric oxide synthase gene polymorphism with major depression and fluoxetine response. Neuropsychobiology. 2003;47:137–40. doi: 10.1159/000070582. [DOI] [PubMed] [Google Scholar]
- Yu Y-Y, Tsai S-J, Chen T-J, et al. Association study of the serotonin transporter promoter polymorphism and symptomatology and antidepressant response in major depressive disorders. Mol Psychiatry. 2002;7:1115–19. doi: 10.1038/sj.mp.4001141. [DOI] [PubMed] [Google Scholar]
- Yura A, Kiuchi Y, Uchikawa T, et al. Possible involvement of calmodulin-dependent kinases in Ca2++dependent enhancement of [H3]5-hydroxytryptamine uptake in rat cortex. Brain Res. 1996;738:96–102. doi: 10.1016/0006-8993(96)00762-7. [DOI] [PubMed] [Google Scholar]
- Zanardi R, Benedetti F, Di Bella D, et al. Efficacy of paroxetine in depression is influenced by a functional polymorphism within the promoter of the serotonin transporter gene. J Clin Psychopharmacol. 2000;20:105–6. doi: 10.1097/00004714-200002000-00021. [DOI] [PubMed] [Google Scholar]
- Zhang HT, Huang Y, Jin SL, et al. Antidepressant-like profile and reduced sensitivity to rolipram in mice deficient in the PDE4D phosphodiesterase enzyme. Neuropsychopharmacology. 2002;27:587–92. doi: 10.1016/S0893-133X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang H, O’Donnell JM. Antidepressant-induced increase in high-affinity rolipram binding sites in rat brain: dependence on noradrenergic and serotonergic function. J Pharmacol Exp Therap. 2003;307:246–53. doi: 10.1124/jpet.103.053215. [DOI] [PubMed] [Google Scholar]
- Zill P, Baghai TC, Zwanzger P, et al. Association analysis of a polymorphism in the G-protein stimulatory alpha subunit in patients with major depression. Am J Med Genet. 2002;114:530–2. doi: 10.1002/ajmg.10409. [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB, Maher BS, 3rd, et al. Genetic linkage of region containing the CREB1 gene to depressive disorders in women from families with recurrent, early-onset, major depression. Am J Med Genet. 2002;114:980–7. doi: 10.1002/ajmg.b.10933. [DOI] [PubMed] [Google Scholar]