Figure 4.
Abnormal ISCU Transcripts and Predicted Alteration of the Peptide Sequence
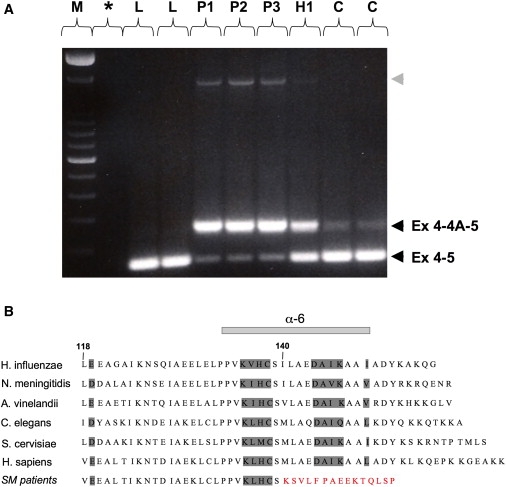
(A) RT-PCR between exons 4 and 5, showing two different transcripts in patients and controls, the normal transcript containing exons 4 and 5, and one containing exons 4, 4A, and 5, as confirmed by cDNA sequencing. The retention of exon 4A is markedly enhanced in patient muscle. Both transcripts are present in the heterozygote. Note the presence of an additional larger transcript in patient muscle (gray arrowhead), for which cDNA sequencing revealed the presence of exons 4, 4A, and 5, as well as the retention of the intronic sequence between exon 4A and exon 5. These transcripts are in very low abundance, and their rare presence in control muscle as well (data not shown) strongly argues against their having a role in the disease. L indicates cDNA from lymphoblastoid cells from controls; P, H, and C indicate cDNA from muscle of the three northern Swedish patients, the heterozygote, and controls, respectively; M indicates 100 bp DNA ladder (Invitrogen); and ∗ indicates no cDNA control.
(B) Alignment of residues encoded by exons 4 and 5, showing the strong conservation of ISCU among species, as well as the alteration of the C terminus of ISCU resulting from the splice mutation in patients (SM). Numbers designate ISCU protein-codon residues, and the residues corresponding to the α6-helix are indicated.
