Figure 2.
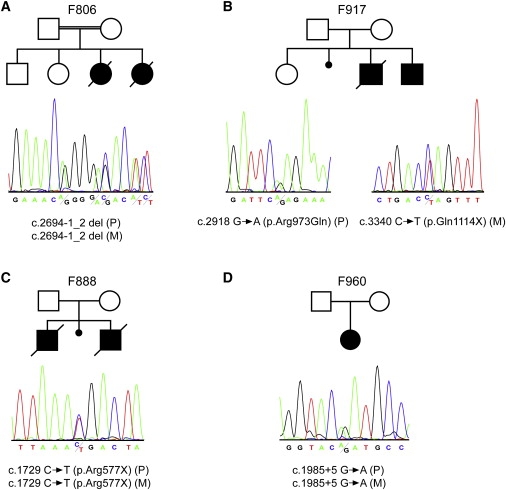
NPHP3 Mutations in Four Families
The upper panel shows the pedigree structure of the families, and the lower panel shows sequence chromatograms illustrating the predicted NPHP3 loss-of-function mutations in the described families. Mutations segregated with the disease status in each family (P indicates paternally inherited allele; M indicates maternally inherited allele). In case of a homozygous mutation, a heterozygous electropherogram of one of the parents is shown for reasons of clarity.
(A) Sequence chromatogram depicting the deletion of the canonical acceptor splice site nucleotides at position −1_2 of the intron 19 splice acceptor site predicted to result in out-of-frame transcripts and a premature stop.
(B) On the left, a sequence chromatograph of the affected individual of family 917 showing a heterozygous G → A exchange which predicts an p.Arg973Gln amino acid substitution is displayed. On the right, a heterozygous C → T exchange predicting a stop codon (p.Gln1114X) is shown.
(C) Sequence chromatograph showing a heterozygous C → T exchange that predicts a stop codon (p.Arg577X).
(D) Sequence chromatogram illustrating the heterozygous G → A exchange at position +5 of the intron 13 splice donor site resulting in skipping of exon 13 and an out-of-frame transcript that leads to premature termination of translation (Figure S2).
