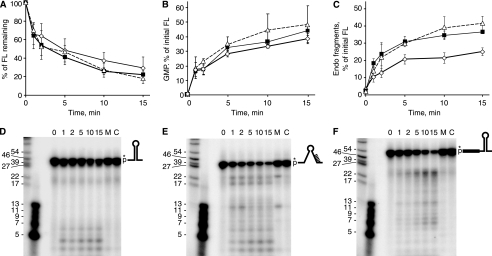
FIGURE 5.
Analysis of RNase J1 activity on RNA oligonucleotides representing 3′-terminal RNA fragments. A–C, data from an 8% denaturing polyacrylamide gel (A), from thin layer chromatography (B), and from a 22.5% denaturing polyacrylamide gel (C) were for 3′-stem RNA (open diamonds), 3′-no-stem RNA (filled squares), and 3′-stem+5′-ext RNA (open triangles). D–F, primary data from 22.5% denaturing polyacrylamide gels for the three substrates, as indicated by the schematic diagrams. Control lanes C, RNA substrate incubated without RNase J1. Lanes M, RNA substrate incubated with mutant RNase J1. Very small size markers (second lane from left) were 5′-end-labeled DNA oligonucleotides of the indicated sizes. Data are the average of four experiments ± S.D. FL, full length.