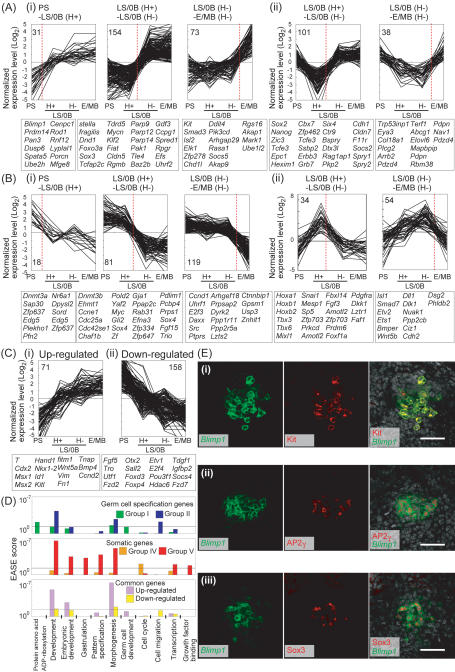
Figure 3.
Complex expression dynamics of the specification and somatic genes. (A–C) Expression dynamics of the specification genes (A), somatic genes (B), and genes commonly regulated in the E/MB stage PGCs and their somatic neighbors (C). The specification and somatic genes are classified based on their dynamics: group I (A, panel i), group II (A, panel ii), group IV (B, panel i), and group V (B, panel ii) (see text for details). Each group is subclassified based on the developmental intervals where the genes’ greatest expression level changes are observed (red dashed line): Intervals between the PS-stage and LS/0B-stage Hoxb1-negative cells, between the LS/0B-stage Hoxb1-positive and Hoxb1-negative cells, and between the LS/0B-stage Hoxb1-positive and E/MB-stage PGCs. Normalized log2 expression levels are plotted. Numbers of the genes that belong to each subclass and representative genes are shown. (D) Functional annotation of the genes over-represented in the indicated groups. EASE scores (see the Materials and Methods) are represented with bars. (E) Immunofluorescence analysis of Kit (panel i), AP2γ (panel ii), and Sox3 (panel iii) in Blimp1-mVenus wild-type embryos at the MB stage. Merged images with DAPI are shown in the right panel. Bar, 50 μm.