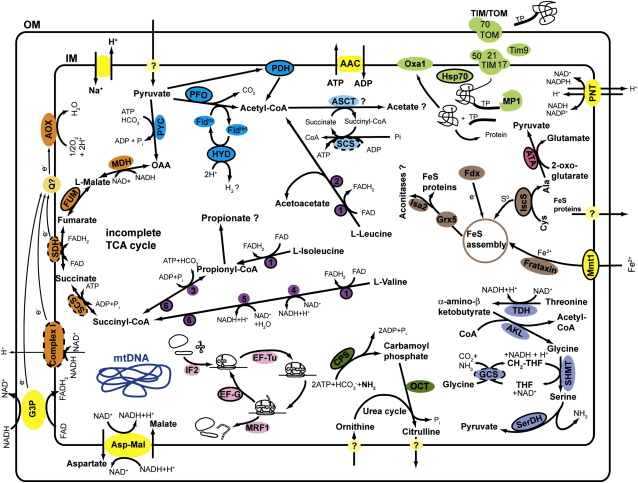
Figure 3.
Proposed Metabolic Map of the Blastocystis Mitochondrion-like Organelles
Proteins with predicted leader peptides have solid black outlines and protein complexes for which leader peptides for only some of the subunits have been predicted have dashed black outlines. The blue pathway represents the conversion of pyruvate to acetyl-CoA via the pyruvate dehydrogenase complex (PDH) or pyruvate:ferredoxin oxidoreductase (PFO). PFO reduces flavodoxin (Fld) and the [FeFe] hydrogenase (HYD) oxidizes flavodoxin with the concomitant production of molecular hydrogen; the question mark indicates that hydrogen production has not been demonstrated for Blastocystis. Acetyl-CoA is converted by the acetate:succinate CoA transferase (ASCT) to acetate (acetate production has not been tested for Blastocystis, but we identified an acetyl-CoA hydrolase like that known to function as ASCT in Trichomonas hydrogenosomes [35]). Transporters and translocators are shown in yellow. Components of the ETC and the partial TCA cycle are orange. Proteins involved in DNA transcription and translation are light pink. The mitochondrial part of the urea cycle is green. Pathways for the metabolism of the amino acids leucine, valine, and isoleucine are purple. Protein import and folding are light green. Pathways of the iron-sulfur cluster assembly are depicted in brown. Light blue represents the glycine-cleavage-system pathway. The question mark next to propionate shows that propionate production has not been assayed for Blastocystis; propionate is a metabolic end product of succinate degradation. For a complete list of abbreviations and numbers used in this figure see the Supplemental Data.