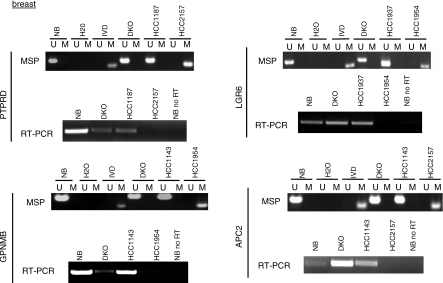
Figure 2. Common Gene Targets of Mutation and Hypermethylation in Breast Cancer.
MSP and RT-PCR expression analysis of selected genes. Each set of MSP and expression analyses are labeled with the corresponding gene name. U denotes the unmethylated band, and M denotes the methylated band. In vitro methylated DNA (IVD) was used as a positive control for methylation. cDNA from normal breast (NB) and normal colon (NC) (Figure 3) was used to determine the expression of the genes in normal tissue. DKO corresponds to DNMT 1/3b double knockout HCT116 cells. All RT-PCR experiments were performed in parallel without reverse transcriptase as a control, and in all cases, no PCR product was generated (unpublished data). The control experiment performed without RT for normal breast is shown (NB no RT) for each gene. RT-PCR was performed for all cancer lines with beta-actin primers to control for the amount of cDNA (Figure 3, bottom).