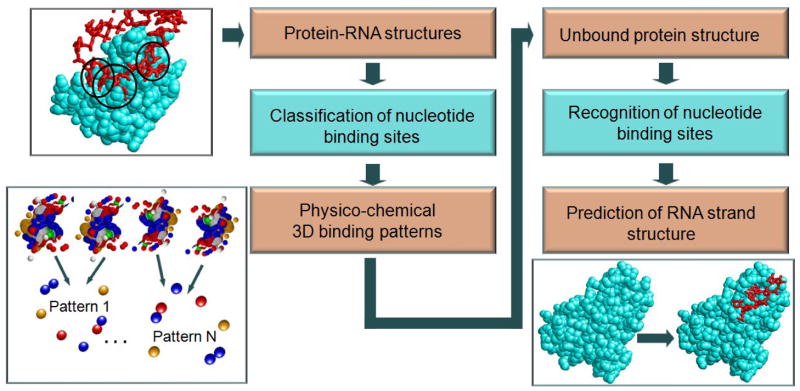
Figure 2. The algorithmic framework.
The framework consists of two main stages. First, we classify all known nucleotide and dinucleotide binding sites and recognize the common types of the physico-chemical binding patterns (left flowchart). Then, we use these patterns for the prediction of dinucleotide binding sites and for the reconstruction of RNA fragments and protein-RNA complexes (right flowchart).